Sponge-based environmental DNA detection as a useful tool in monitoring Mycobacterium tube
May 27, 2025
Abstract
The European bison (Bison bonasus), also called wisent, is the largest terrestrial mammal in Europe, classified by the International Union for Conservation of Nature (IUCN) as a “Near Threatened� species. Tuberculosis (TB) represents a well-known threat to wisent, especially nowadays when infectious diseases are emerging to this species, due to locally high population density and frequent translocation (and consequently increased exposure to infectious diseases). There is an urgent need to control the TB-epidemiological situation in the European bison environment. This study aimed to assess the usefulness of the sponge-based environmental-DNA (eDNA) for monitoring TB in free-ranging and captive European bison herds based on the knowledge of the TB-epidemiological situation in the past. Between 2022 and 2024, eDNA samples (n = 84) were collected from European bison or their environment in eight herds from different regions of Poland. The real-time PCR techniques with IS6110, IS1081, and MPB70 as targets were used to detect MTC DNA markers in the samples. The MTC DNA IS6110 and IS1081 were simultaneously detected in 17/84 (20.2%) samples. No sample was positive for MPB70. The highest number of positive results for both markers (IS6110 and IS10081 targets) was in the captive herd in Bieszczady-Muczne, followed by the free-ranging herd in the nearby Bieszczady Mountains. Even though detecting nucleic acid, especially at low eDNA signal, does not necessarily indicate viable pathogens, our results suggest this new approach could represent a suitable complementary tool for TBC surveillance in wildlife-livestock interface of particular interest in endangered species.
Introduction
Animal tuberculosis (TB) is a chronic infection of cattle and many other domestic and wild mammals. TB leads to a loss in productivity in livestock and significant economic costs1,2. This zoonotic disease is still a crucial public health problem3. Although many countries have made costly efforts to eliminate the disease, eradication remains elusive. TB also remains a serious threat to wildlife conservation strategies and biodiversity4,5. One of the major problems for effective monitoring, is the well-known set of limitations of existing diagnostic tests5, this situation is particularly worrying in wild animals, such as European bison (Bison bonasus), called also a wisent, which is the largest land mammal in Europe.
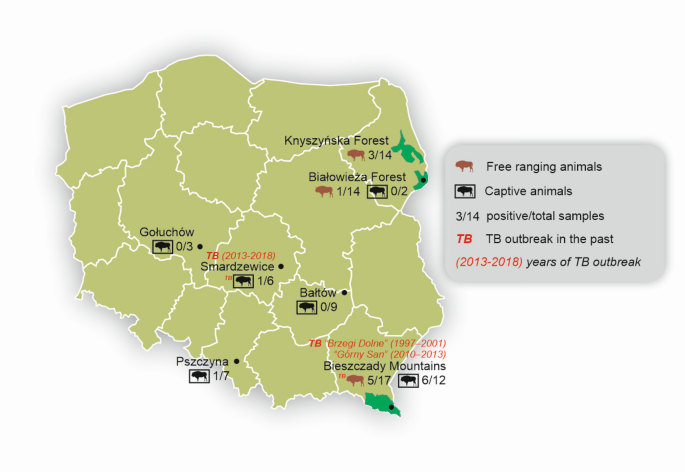
Although Poland remains an officially tuberculosis-free (OTF) country, the causative agents of TB have been isolated in wildlife in Poland, including wild boar (Sus scrofa), wolves (Canis lupus) and European bison (Bison bonasus)6,7,8,9. The European bison seems to be a particularly sensitive species10. Its status is defined by the International Union for Conservation of Nature (IUCN) as “Near Threatened�11. Currently, one of the main threats to this species is the excessive local density of the population (e.g12). Another relevant epidemiological risk factor is the frequent captive breeding and translocation13. In 1996–2013, 45 cases of TB in European bison (Bison bonasus caucasicus) were microbiologically confirmed in the Bieszczady Mountains in southeastern Poland14. The Bieszczady Mountains are not the only region where TB has occurred in free-ranging European bison. In spring 2016, in the Borecka Forest, two cases of TB were found in an 8-year-old and a 13-year-old bull, respectively6. However, no positive results were confirmed in this region afterwards. In the last five years, during the TB monitoring in European bison, there have been positive results in gamma-interferon assays in Knyszyńska Forest but there was no confirmation by culture in material collected post-mortem (unpublished data).
TB also occurred in captive European bison herds. At the end of 2013, there were 21 individuals in the European Bison Breeding Centre in Smardzewice (central Poland). Since that year, the herd was completely depopulated in 2018 and Mycobacterium caprae infection was confirmed in all animals15. The study revealed the most likely source of infection was European bison from the Silesian Zoo (in which 11 animals, including 3 giraffes, 5 antelopes and 1 alpaca were found TB infected in 2009–2010)16,17. In 2013, two bulls were transported from Smardzewice to the “Wolisko� enclosure in the Borecka Forest. Due to the possibility of transferring the infection from Smardzewice, in September 2013, the animals were depopulated, and in one of them, TB was confirmed18. In 2024, two PCR-positive results (unpublished data) were confirmed in nasal swabs collected during the immobilization of European bison from two captive herds: Bałtów (two animals) and Gołuchów (one animal), as well as two doubtful gamma-interferon test (IGRA) results in Bałtów (unpublished data).
TB remains one of the major health threats to endangered European bison19 although the current TB situation in this species remains unclear. Ante-mortem diagnosis of TB by tuberculin skin testing is expensive because it requires a double immobilization of animals, which is associated with risks to their health20. Even though there have been attempts to develop improved diagnostic methods for tuberculosis in European bison15,21,22, results are often ambiguous implying a limited number of detection TB tools for European bison is currently available. An important direction for improving TB diagnostics is to create a diagnostic algorithm that includes the inclusion of several tests to achieve overall high sensitivity and specificity. Another important step involves new technologies like testing environmental DNA (eDNA)23. This latter is particularly important in monitoring wildlife and the wildlife-livestock interface as it allows Mycobacterium tuberculosis complex (MTC) eDNA monitoring and eDNA based risk analyses24,25. In fact in these scenarios, improving a diagnostic method should be aimed at developing tools to identify the pathogen in all hosts and settings and not specific to a certain host type, which furthermore highlights the role and importance of eDNA-based methods. The importance of certain environmental features in maintaining TB infection in multi-host systems has been confirmed due to long-lasting MTC persistence in the environment26,27.
In this study, we revisited seven of the European bison populations studied in a recent serosurvey in Poland21 implementing environmental nucleic acid detection (ENAD) using non-invasive samplings for molecular monitoring of MTC markers in free-ranging and captive European bison herds.
Results
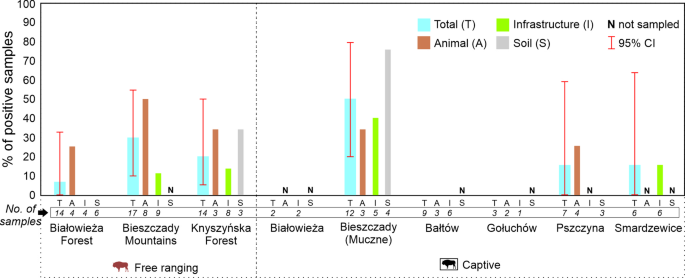
The IS6110 and IS1081 markers were simultaneously detected by PCR in 17/84 (20.2%; 95%CI: 12.86–30.27) samples (Ct range 35.25 to 37.81 for IS6110, and 38.15 to 39.64 for IS10081). No sample was positive for MPB70. The highest number of positive results was observed in the captive herd in Bieszczad-Muczne, followed by the nearby free-ranging herd in the Bieszczady Mountains (Fig. 1). Overall, IS6110 and IS1081 were detected in sponge samples collected from animals: 8/27 (29.6%; 95%CI: 12.86–30.27), feeders: 5/34 (14.7%; 95%CI: 5.98–30.65), and soil: 4/16 (37.5%, 95%CI: 17.78–62.83).
Both the TB outbreak in European bison in the past and the source of samples had a significant impact on MTC DNA markers detection in sponge samples (Table 1). Animals in free-ranging or captive locations with a previous TB outbreak showed a significantly higher frequency of MTC DNA markers in samples (p < 0.004). The source of samples was included in the model, however, did not statistically influence the level of MTC DNA markers frequency detection (Table 1). The percentage of positive samples obtained with antibodies against P22 (MTC) in European bison21 and MTC DNA (IS6110 and IS1081 targets) obtained in this study were highly correlated (Rho = 0.832, p = 0.020).
Discussion
In protected wildlife species, sample collection for disease surveillance purposes is extremely difficult. Finding reliable, non-invasive methods is crucial to detect and monitor infectious diseases and to eventually assess the impact of disease control interventions. In the present study, we have demonstrated the usefulness of sponge-based non-invasive sampling to detect MTC markers in the European bison environment, an endangered species susceptible to TB. Previous studies have demonstrated the ability of this approach to detect MTC DNA24,25 and other pathogens25,28,29 not only in domestic environments but also in natural settings28, even at the livestock-wildlife interface24,25,30.
In our study we have found a strong link between the existence of TB records in European bison herds in the past and the probability of IS6110 and IS1081 detection. Furthermore, our results are in line with previous detection of P22 antibodies in European bison in the tested regions. A high correlation of results in those two independent methods (serology and eDNA detections) strengthens their credibility.
The highest number of positive results were detected in Bieszczady in both captive (Bieszczady-Muczne) and free-ranging herds (Bieszczady Mountains). This is in line with the region’s TB history9 as long as the two biggest TB outbreaks in European bison were reported in this part of Poland14. In fact, in the last five years, there have been confirmed TB cases in other wildlife species in this region: in the brown bear (Ursus arctos) (MTC DNA detection)31, wild boar, and wolves (culture)32 which confirms the suitability of non-invasive sampling to detect the circulation of MTC in close environments to European bison. Due to the high number of positive results in a captive herd in Bieszczady-Muczne detected in this study, active surveillance should be implemented in the region due to the potential impact on health as well as biodiversity.
Another site where IS6110 and IS1081 targets were detected was a captive European bison herd in Smardzewice. A TB outbreak occurred there between 2013 and 2018 (the entire herd was culled in 2018). Since then, there have been no European bison in this area. However, it cannot be excluded that other species could act as TB reservoirs like that reported in wild boar in the Bieszczady Mountains7.
In Knyszyńska Forest, IS6110 and IS1081 positive samples were also obtained. No TB cases have been confirmed in European bison or other wildlife species in the region. However, P22 antibodies (10.8%) and positive gamma-interferon results have recently been obtained in animals of the region [Didkowska, unpublished data]21.
Even though detecting nucleic acid, especially at low DNA signal (the lowest Ct value in this study was 35.25 in a European bison surface from Bieszczady-Muczne), does not necessarily indicate viable pathogens in the environment, it may indicate the circulation of MTC or related bacteria. Our results indicate that non-invasive samplings could represent a convenient tool to detect regions in which surveillance efforts should be reinforced.
Our study has some limitations. Firstly, DNA-based methods do not distinguish between viable and dead MTC or related bacteria which represents eDNA detection could not reflect current risk33. Finding MTC markers eDNA in sponge samples, including in those collected from animals, does not confirm the infection at the individual level but may inform about epidemiological connection with other herd members, other wildlife species, or livestock, especially in studies in free-living herds24. In fact, in our study, the TB-history of each tested region of Poland, P22 serology, and/or MTC bacteriological results have shown that eDNA detection may reflect the MTC situation of a region providing the opportunity of countless samplings (minimizing the contact with animals) to better understand the pattern of IS6110 and IS1081 positivity in the studied European bison herds. Another limitation of our study that could be considered is the lack of information on TB status (at individual and herd levels) in parallel with environmental samplings could be considered. However, the aim of this study was to evaluate the suitability of a non-invasive approach for MTC markers monitorization in European bison (not to establish the current TB epidemiological situation). Another limitation to consider is that only IS6110 and IS1081 positive results were obtained and MPB70 were negative. However, the obtained Ct values were rather high, and this could explain why a multicopy target (IS6110 and IS1081) was detected and a single unique copy (MPB70) was not detected. Besides, IS6110 can also be present in other organisms and therefore we claim marker detection rather than MTC detection. However, we note that our findings based on IS6110 and IS1081 markers make biological sense, as explained above.
We suggest that sponge-based eDNA sampling provides a good complementary detection tool to monitor MTC markers and probably other pathogens in the wild-environment to implement disease surveillance in wildlife species such as European bison.
Materials and methods
Samplings
Between 2022 and 2024, sponges for eDNA MTC detection (n = 84) were collected from European bison or their environment in three free-ranging and six captive populations. In Poland (Table 2; Fig. 2). In the case of free-ranging herds, sampling surfaces were selected based on knowledge about the presence of European bison in each area or data from camera traps.
Mycobacterium tuberculosis complex DNA (IS6110 and IS1081 targets) detection in sponge samples collected from European bison (Bison bonasus) or its environment by maintenance type (free ranging or captive) and site (six different regions of Poland). The figure was generated in Quantum GIS v.3.4.5 (https://qgis.org) and CorelDRAW Standard 2020 (https://www.corel.com). One of the coauthors created the map.
Sponges were collected from animals during anaesthesia for routine diagnosis before transport (n = 8) or immediately after culling (n = 19), and from abiotic surfaces including feeders, fences, soil, and trees (n = 57) (Table 3). The sampling procedure followed manufacturer instructions (GPSponge® KIT, Genetic PCR Solutions, Alicante, Spain). Briefly, a sponge prehydrated with 15 ml of an isotonic surfactant liquid was scrubbed by passing over each location ten times24.
DNA extraction
The extraction and purification of DNA from the liquid of the sponge samples was performed using the QIAamp Fast DNA Stool Mini Kit (Qiagen Hilden, Germany), starting from the pellet obtained after centrifuging 900 µL of the previous homogenized sample for 3 min at 13.000 rpm.
PCR for MTC detection
The real-time PCR with IS6110, IS1081, and MPB70 as targets were used to detect MTC in the samples, using the primers described previously34,35,36,37. We defined three real-time PCR targets to detect MTC DNA in environmental samples, IS6110, IS1081, and MBP70. The primer sequences for the IS6110 multicopy target were 6110-forward: 5´- GGTAGCAGACCTCACCTATGTGT-3´, 6110-reverse: 5´- AGGCGTCGGTGACAAAGG-3´ and 6110-probe: 5´- FAM- CACGTAGGC GAACCC – MGB NFQ-3´35. The primer sequences for the MPB70 monocopy target were mpb70 – forward: 5′-CTCAATCCGCAAGTAAACC-3′, mpb70-reverse: 5′-TCAGCAGTGACGAATTGG-3′ and mpb70-probe: 5′- FAM CTCAACAGCGGTCAGTACACGGT-BHQ1-3′34. The primers sequences for IS1081 were TR IS1081 F- forward: 5´CCG CCA CCG TGA TTT CGA3´, TR IS1081 R-reverse: 5´GCC AGT CCG GGA AAT AGC T 3´ and TR IS1081 P-probe: (FAM)- 5´CCG CAA CCA TCG ACG TC- 3´-(MGB-NFQ)37.
PCR was performed using the QuantiFast Pathogen + IC Kits (400) according to the manufacturer’s instructions (QIAGEN). This kit includes an internal amplification control (IAC), as well as specific reagents and primers/probes required for amplification. Ultra-pure distilled water was used as the negative control and DNA from a BCG inoculum was used as the positive control. All IS6110 and MPB70 PCR reactions were performed in a CFX96 TouchTM real-time PCR detection system (Bio-Rad, Hercules, CA, USA) under the following cycling conditions: 95ºC for 5 min, followed by 45 2-step cycles of 95ºC for 15 s and 60ºC for 1 min.
The thermal cycler protocol for the IS1081 PCR were as follows: an initial activation of Taq polymerase at 95 °C for 5 min, followed by 40 cycles (denaturation at 95 °C for 15 s + annealing/extension at 60 °C for 30 s).
Samples were considered positive if, at least, positive results were obtained for IS6110 (Ct value ≤ 38) and IS1081 (Ct ≤ 40).
Statistical analyses
The probability of finding MTC eDNA markers (IS6110 and IS1081 targets simultaneously) was verified by the presence in samples with the generalized linear binary model in IBM SPSS software. Positive vs. negative MTC eDNA markers samples (IS6110 and IS1081) (marked as 1 and 0 respectively) were tested as dependent variables regarding previous TB outbreaks (Bieszczady Mountains – free-ranging animals and captive herds in Muczne and Smardzewice) and source of samples. The source of samples was in three groups: animals, elements (feeder, trees, fence), and soil. Then, we performed a pairwise comparison of marginal means with the LSD test38.
The relation between the percent of positive samples obtained with antibodies against P22 (MTC) in European bison21 and MTC markers DNA (IS6110 and IS1081 targets simultaneously) obtained in this study was verified. For this purpose, Spearman’s rank correlation coefficient in IBM SPSS (version 29.0) was used.
Data availability
All detailed data are available for request in the corresponding authors.
References
-
Olmstead, A. L. & Rhode, P. W. An impossible undertaking: the eradication of bovine tuberculosis in the united States. J. Econ. Hist. 64, 734–772 (2004).
-
Palmer, M. V. & Waters, W. R. Bovine tuberculosis and the establishment of an eradication program in the United States: role of veterinarians. Vet. Med. Int. 2816345, (2011). https://doi.org/10.4061/2011/816345 (2011).
-
Olea-Popelka, F. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis—a call for action. Lancet Infect. Dis. 17, 21–25. https://doi.org/10.1016/S1473-3099(16)30139-6 (2017).
-
Gortázar, C., Che Amat, A. & O’brien, D. J. Open questions and recent advances in the control of a multi-host infectious disease: animal tuberculosis. Mammal Rev. 45, 160–175 (2015).
-
Thomas, J., Balseiro, A., Gortázar, C. & Risalde, M. A. Diagnosis of tuberculosis in wildlife: a systematic review. Vet. Res. 52, 31 (2021).
-
Krajewska-Wędzina, M. et al. Ten Years of Animal Tuberculosis Monitoring in Free-Living European Bison (Bison bonasus) in Poland. Animals, 13, 1205. (2023). https://doi.org/10.3390/ani13071205
-
Welz, M. et al. The eradication of M. Caprae tuberculosis in wild Boar (Sus Scrofa) in the Bieszczady mountains, Southern Poland – An administrative perspective. J. Vet. Res. 67, 61–66 (2023).
-
Orłowska, B. et al. Mycobacterium caprae transmission to free-living grey wolves (Canis lupus) in the Bieszczady mountains in Southern Poland. Europ J. Wildl. Res. 63 (5), 1. https://doi.org/10.1007/s10344-017-1079-4 (2017).
-
Orłowska, B. et al. Epidemiological characterization of Mycobacterium caprae strains isolated from wildlife in the Bieszczady mountains, on the border of Southeast Poland. BMC Vet. Res. 16(1), 362. https://doi.org/10.1186/s12917-020-02581-3 (2020).
-
Didkowska, A. et al. Microbiological and molecular monitoring for bovine tuberculosis in the Polish population of European bison (Bison bonasus). AAEM 28, 575–578. https://doi.org/10.26444/aaem/130822 (2021).
-
Plumb, G., Kowalczyk, R. & Hernandez-Blanco, J. A. Bison bonasus. The IUCN Red List of Threatened Species (2020).
-
Sobczuk, M. & Olech, W. Damage to the crops inflicted by European bison living in the Knyszyn forest. Eur. Bison Conserv. Newsl. 9, 39–48 (2016).
-
Klich, D., Olech, W., �opucki, R. & Danik, K. Community attitudes to the European bison Bison Bonasus in areas where its reintroduction is planned and in areas with existing populations in Northeastern Poland. Eur. J. Wildl. Res. 64, 61 (2018).
-
Krajewska, M. et al. Transmission of Mycobacterium caprae in a herd of European bison in the Bieszczady mountains, Southern Poland. Eur. J. Wildl. Res. 61, 429–433 (2015).
-
Didkowska, A. et al. Biopsy and Tracheobronchial Aspirates as Additional Tools for the Diagnosis of Bovine Tuberculosis in Living European Bison (Bison bonasus). Animals 10, (2017). https://doi.org/10.3390/ani10112017, (2020).
-
Augustynowicz-Kopeć, E., Krajewska, M., Zabost, A., Napiórkowska, A. & Zwolska, Z. Characterisation of Mycobacterium bovis strains isolated from farm and wild animals from Poland. Bull. Vet. Inst. Pulawy. 55, 381–383 (2011).
-
Krajewska-Wędzina, M., Augustynowicz-Kopeć, E., Weiner, M. & Szulowski, K. Treatment for active tuberculosis in giraffe (Giraffa camelopardalis) in a zoo and potential consequences for public health – Case report. Ann. Agr Env Med. 25, 593–595 (2018).
-
Bielecki, W. et al. Monitoring Zdrowia Populacji Żubrów Jako element ochrony Gatunku. Eur. Bison Conserv. Newsl. 7, 43–50 (2014). (2014).
-
Larska, M. & Krzysiak, M. K. Infectious disease monitoring of European bison (Bison bonasus). In wildlife population monitoring. IntechOpen https://doi.org/10.5772/intechopen.84290 (2019).
-
Gardoni, N. et al. Arterial oxygenation and acid-base status before and during oxygen supplementation in captive European bison (Bison bonasus) immobilized with etorphine-acepromazine-xylazine. Front. Vet. Sci. 10 https://doi.org/10.3389/fvets.2023.112591 (2023).
-
Didkowska, A. et al. Antibodies against the Mycobacterium tuberculosis complex and Brucella spp. In captive and free-living European bison (Bison bonasus) In Poland. Vet. Med. Sci. 10, e1314. https://doi.org/10.1002/vms3.1314 (2024).
-
Didkowska, A. et al. Antibody responses in European bison (Bison bonasus) naturally infected with Mycobacterium caprae. Vet. Microb. 253 https://doi.org/10.1016/j.vetmic.2020.108952 (2021).
-
Gortázar, C., de la Fuente, J., Perelló, A. & DomÃnguez, L. Will we ever eradicate animal tuberculosis? Ir. Vet. J. 76 https://doi.org/10.1186/s13620-023-00254-9 (2023).
-
MartÃnez-Guijosa, J. et al. Environmental DNA: a promising factor for tuberculosis risk assessment in multi-host settings. PLoS ONE. 15 https://doi.org/10.1371/journal.pone.0233837 (2020).
-
Herrero-GarcÃa, G. et al. One health farming: noninvasive monitoring reveals links between farm vertebrate richness and pathogen markers in outdoor hoofstock. One Health. 19, 100924. https://doi.org/10.1016/j.onehlt.2024.100924 (2024).
-
Barasona, J. A. et al. Environmental presence of Mycobacterium tuberculosis complex in aggregation points at the wildlife/livestock interface. Transbound. Emerg. Dis. 64 (4), 1148–1158. https://doi.org/10.1111/tbed.12480 (2017).
-
Hopkins, S. R. et al. Environmental Persistence of the World’s Most Burdensome Infectious and Parasitic Diseases. Front. Public. Health. 10 https://doi.org/10.3389/fpubh.2022.892366 (2022).
-
Fernández-de-Mera, I. G. et al. Detection of environmental SARS-CoV-2 RNA in a high prevalence setting in Spain. Transboun Emerg. Dis. 68, 1487–1492. https://doi.org/10.1111/tbed.13817 (2021).
-
Rebollada-Merino, A. et al. Environment and offspring surveillance in Porcine brucellosis. Front. Vet. Sci. 9 https://doi.org/10.3389/fvets.2022.915692 (2022).
-
Herrero-GarcÃa, G. et al. Non-invasive surveillance of shared pathogens in the Eurasian brown bear (Ursus arctos) human interface. One Health. 18 https://doi.org/10.1016/j.onehlt.2024.100746 (2024).
-
Orłowska, B. et al. Detection of Mycobacterium tuberculosis complex genetic material in a Free-Living brown bear (Ursus arctos). J. Wild Dis. 59, 539–541. https://doi.org/10.7589/JWD-D-22-00150 (2023).
-
Radulski, Å�. Wykrywanie zakażeÅ„ bakteriami z rodzaju Mycobacterium u zwierzÄ…t wolno żyjÄ…cych i towarzyszÄ…cych przy użyciu nowoczesnych metod badawczych. PhD Thesis, PaÅ„stwowy Instytut Weterynaryjny – PaÅ„stwowy Instytut, Poland. (2022).
-
Pereira, A. C., Pinto, D. & Cunha, M. V. First time whole genome sequencing of Mycobacterium bovis from the environment supports transmission at the animal- environment interface. J. Hazard. Mater. 472 https://doi.org/10.1016/j.jhazmat.2024.134473 (2024).
-
Lorente-Leal, V. et al. Validation of a Real-Time PCR for the detection of Mycobacterium tuberculosis complex members in bovine tissue samples. Front. Vet. Sci. 6 https://doi.org/10.3389/fvets.2019.00061s (2019).
-
Lorente-Leal, V. et al. Direct PCR on tissue samples to detect mycobacterium tuberculosis complex: an alternative to the bacteriological culture. J. Clin. Microbiol. 59 https://doi.org/10.1128/JCM.01404-20 (2021).
-
Michelet, L., de Cruz, K., Karoui, C., Hénault, S. & Boschiroli, M. L. Mycobacterium microti, un agent tuberculeux méconnu In: Barbara Dufour, editor. Épidémiol Et Santé Anim. Maisons-Alfort Cedex, France: Journée scientifique AEEMA, 24 mars 2017, 71, 129–138 (2017).
-
Michelet, L. et al. Second line molecular diagnosis for bovine tuberculosis to improve diagnostic schemes. PLoS One. 13 (11), e0207614. https://doi.org/10.1371/journal.pone.0207614 (2018).
-
Burnham, K. P. & Anderson, D. R. Model Selection and Multimodel Inference: A Practical Information–Theoretic Approach (Springer, 2002).
Acknowledgements
We would like to thank all involved vets, foresters, and zootechnicians for their help in collecting sponges. This work was supported by the Spanish MCIN/AEI/10.130 39/501100011033/the European Union NextGeneration EU/PRTR [grant number PLEC2021-008113] and EU Junta de Comunidades de Castilla-La Mancha [grant number SBPLY/23/180225/000008]. The publication was financed by the Science Development Fund of the Warsaw University of Life Sciences – SGGW.
Funding
This work was supported by the Spanish MCIN/AEI/10.130 39/501100011033/the European Union NextGeneration EU/PRTR [grant number PLEC2021-008113] and EU Junta de Comunidades de Castilla-La Mancha [grant number SBPLY/23/180225/000008]. The publication was financed by the Science Development Fund of the Warsaw University of Life Sciences – SGGW.
Author information
Authors and Affiliations
Contributions
AD research design and coordination, collecting samples, data analysis, writing – original draft; MP-S, CH – laboratory work, revision of the manuscript, DK data analysis, statistical analysis, revision of the manuscript, KA, LW, LD data analysis, revision of the manuscript, CG research design and coordination, data analysis, revision of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Ethical approval
All experimental protocols (including culling of European bison) were approved by the General Directorate for Environmental Protection in Poland, based on the Act of 16 April 2004 on The Protection of Nature. All methods were carried out in accordance with relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Didkowska, A., Pérez-Sancho, M., Herranz, C. et al. Sponge-based environmental DNA detection as a useful tool in monitoring Mycobacterium tuberculosis complex markers in European bison (Bison bonasus).
Sci Rep 15, 18503 (2025). https://doi.org/10.1038/s41598-025-01966-4
-
Received: 25 February 2025
-
Accepted: 09 May 2025
-
Published: 27 May 2025
-
DOI: https://doi.org/10.1038/s41598-025-01966-4
Keywords
Search
RECENT PRESS RELEASES
Related Post