Utilizing aquatic environmental DNA to address global biodiversity targets
April 27, 2025
Abstract
Achieving global biodiversity goals requires assessing, attributing and reversing the ongoing, unprecedented biodiversity decline in aquatic ecosystems, and relies on adequate data to inform policy and action. Analysis of environmental DNA (eDNA) has become established as a novel and powerful approach to assess the state and functioning of aquatic ecosystems, and although increasingly implemented by stakeholders its potential is not yet fully tapped. In this Perspective, we review the current state of aquatic eDNA research, focusing in particular on the policy relevance of eDNA and its utility in contributing towards the Kunming–Montreal Global Biodiversity Framework. We summarize key technological developments in eDNA science to measure organismal diversity, its potential for spatial and temporal upscaling to become a key reference for local to global biodiversity action, and the next steps needed to effectively implement eDNA for decision-making and reaching biodiversity targets. Using eDNA to support biodiversity assessment will particularly benefit the understanding of understudied ecosystems and allow the direct calculation of ecological indices and implementation of FAIR (findable, accessible, interoperable and reusable) and inclusive data curation. Important next steps for eDNA require proper method standardization and commonly agreed quality standards, populating reference databases, and overcoming methodological constraints in retrofitting novel eDNA-based approaches to existing biodiversity monitoring approaches.
Key points
-
Aquatic biodiversity is declining from local to global scales, yet in most regions, no or only minimal data on state and change of biodiversity are available.
-
Representative, scalable and replicable monitoring of aquatic biodiversity is needed to achieve the Kunming–Montreal Global Biodiversity Framework targets.
-
Environmental DNA (eDNA) analysis is a key technology to achieve a global measurement network of state and trends in biodiversity, and many of its technical aspects are ready to be implemented.
-
eDNA analysis allows whole-community assessments, broad taxonomic coverage, high spatiotemporal resolution and calculation of environmental indices.
-
Particularly for undersampled regions, large rivers, lakes and marine systems, eDNA metabarcoding might be an effective technology to rapidly gain biodiversity data.
-
To make eDNA-based monitoring policy frameworks successful and trusted, inclusive development and uptake of international method standards are needed.
Similar content being viewed by others
Introduction
The unprecedented decline of global biodiversity in terrestrial, freshwater and marine ecosystems1,2,3, including the loss of functional and genetic diversity, is exceeding the planetary boundary of biosphere integrity4 and threatening many ecosystem functions and services. Consequently, biodiversity loss has been identified as an urgent global challenge5, and increasing political consensus exists that rapid measures to halt and reverse biodiversity loss are needed. The Kunming–Montreal Global Biodiversity Framework (GBF), adopted by the Parties of the Convention on Biological Diversity in December 2022 (ref. 6), sets out four overarching goals for 2050: (1) halt human-induced species extinction, (2) achieve sustainable use of biodiversity, (3) implement equitable sharing of benefits from biodiversity and (4) mobilize resources to close the biodiversity finance gap. The GBF also contains 23 targets to be reached by 2030, including conserving at least 30% of the world’s land and sea areas, restoring degraded ecosystems, reducing pollution, ensuring sustainable use of biodiversity and performing adequate monitoring of biodiversity globally6,7.
Freshwater and coastal marine systems are among the most affected by global change8, and the main drivers of biodiversity decline in these ecosystems are well known9: habitat modification, pollution, invasive alien species, direct exploitation and climate change. Attributing the effects of these large-scale drivers to localized spatial and temporal scales is challenging, primarily because of the transport and mixing of water in aquatic systems. For example, diffuse and point-source inputs of chemicals in rivers have cumulative effects on aquatic biodiversity at a downstream catchment scale10. Furthermore, aquatic ecosystems cross countries, continents and are — via oceans and through the global water cycle — far more continuously integrated than terrestrial systems11. Consequently, effects of anthropogenic pressures have the potential to spread over large spatial extents and across political jurisdictions.
To implement actions that successfully address aquatic biodiversity loss, adequate assessments are necessary7,12 and require monitoring of all aquatic biodiversity — including groundwater, surface water and marine systems. Such monitoring must be broadly applicable, comparable and scalable, and must be implementable for countries worldwide that have different baseline information on species diversity and abundance to effectively inform policymaking.
Environmental DNA (eDNA)13 analysis is a key advance in biodiversity monitoring14 that will help to achieve the GBF targets. eDNA is directly extracted from an environmental sample15, such as water, soil or air (in contrast to DNA directly isolated from specimens) and analysed. Environmental samples might contain the DNA of all organisms present (including microorganisms, meiofauna and macrofauna), and include both intra- and extracellular DNA. eDNA in aquatic systems can also provide information on the diversity of organisms in adjacent terrestrial systems whose DNA is washed into the water bodies16,17. Environmental samples might also contain RNA (eRNA)18,19, which is more difficult to handle, but can reflect more recent and metabolically active community signals than eDNA20,21.
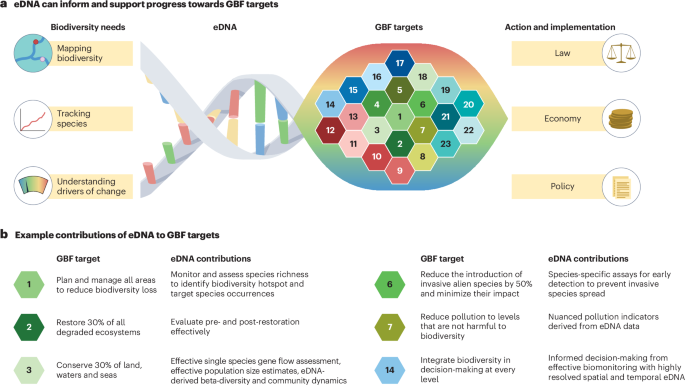
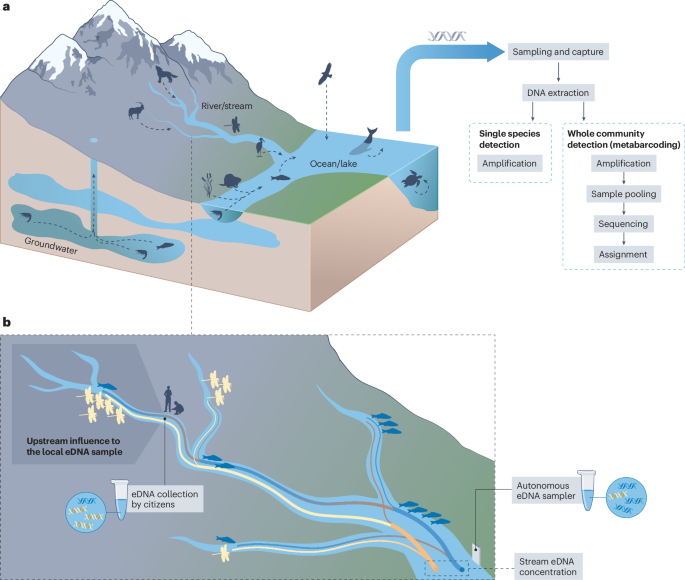
As eDNA can be sampled and analysed across all aquatic ecosystems to detect organisms in a universal manner, it can be applied to meet multiple GBF targets (Fig. 1). Methods based on eDNA are already established and being implemented for the early detection and mitigation of invasive alien species22,23 (GBF target 6: reduce the introduction of invasive alien species by 50%) and the evaluation of the state of ecosystems under multiple stressors and in pre- and post-restoration states24,25 (target 2: restore 30% of all degraded ecosystems). Collecting eDNA samples is becoming increasingly fast and cost-efficient26, rendering this approach highly scalable across space and time. The simplicity of gathering water samples also grants eDNA analysis a high potential for automatization27,28 and for its inclusion in citizen science projects29,30,31. Methods based on eDNA could, therefore, be implemented across the world, even in regions for which we currently lack adequate biodiversity data.
a, The Kunming–Montreal Global Biodiversity Framework (GBF) identified 23 action-oriented global targets to be reached by 2030 (ref. 6). Many of these targets require information on the state, change and trends of biodiversity for scientific, political and economic decision-making. Using environmental DNA (eDNA) to provide information on biological diversity offers a potentially universal approach that can support GBF targets by providing baseline data and action–response information to guide decision-making. Establishing, tracking and assessing biodiversity hotspots, biodiversity trends and change is particularly important. b, Multiple targets can be directly assessed using existing eDNA technologies, which are sufficiently developed for implementation but not yet routinely used in most countries.
In this Perspective, we examine the current state of aquatic eDNA research, with a particular focus on the policy relevance of eDNA and its utility in contributing towards GBF targets (Fig. 1). We focus on eDNA only (not eRNA) because of its higher readiness level for biodiversity monitoring. First, we explain the technical aspects of eDNA-based biodiversity monitoring required for assessing baseline states and action–response effects. Second, we assess the use of eDNA at the level of communities and ecosystems, targeted species, and its ability to attribute environmental change to underlying drivers. Finally, we outline necessary steps, including standardization, data curation and data deposition, to make eDNA analysis globally accessible.
The promise of eDNA
DNA is universal across all species, and traces of DNA are left in the environment by all organisms, for example, by shedding skin cells. Extraction of this DNA from an environmental sample, in theory, enables the detection of any organism, either by targeted screening for single species or assessment of whole communities13,32 (Fig. 2). In both cases15, the first step is sampling water in the focal environment and extracting the DNA present in these samples. For species-specific assays, a portion of DNA is amplified using primers specific to this taxon33,34 and the number of copies present in the original sample is evaluated using a quantitative polymerase chain reaction (qPCR)35 approach, such as real-time qPCR or digital PCR. When assessing whole communities via metabarcoding14, a barcode gene region is amplified using primers with a broader taxonomic scope. The amplified DNA is sequenced via high-throughput sequencing technologies36, which yield millions to billions of sequences (termed ‘reads’). These sequences are then bioinformatically processed and clustered into meaningful units (either as operational taxonomic units or as  amplicon sequence variants) and compared with reference sequences in databases to assign them to specific organisms. These two approaches are well established and are currently the most used methods applied to eDNA samples32,37. New approaches are being developed to investigate the functional or genetic diversity of a group of organisms38, to investigate trophic relationships39 or to obtain information on the whole genome of the organisms present in the samples40,41, but are not yet sufficiently developed for widespread biodiversity monitoring. Generally, sampling of eDNA is applicable to systems with minimal prior information and can also be integrated with data from other monitoring techniques such as remote sensing, camera or audio recordings17,42.
a, Aquatic environmental DNA (eDNA) is sampled from marine, freshwater and groundwater ecosystems, enabling spatial (for example, catchment wide) and temporal (for example, across seasons) integration. The basic principle of aquatic eDNA analysis include sampling and capturing the DNA (usually via filtration) followed by DNA extraction, amplification (either of single species or through metabarcoding), sequencing and taxonomic assignment by comparison to DNA reference databases. b, Environmental DNA collected in river networks enables gathering information on biodiversity from a wider spatial extent by integrating hydrological first principles and estimated production and decay of DNA. Collection of samples can be enhanced via automated methods (such as autonomous samplers) or via citizen science approaches, which enables accumulation of data at high spatiotemporal resolution.
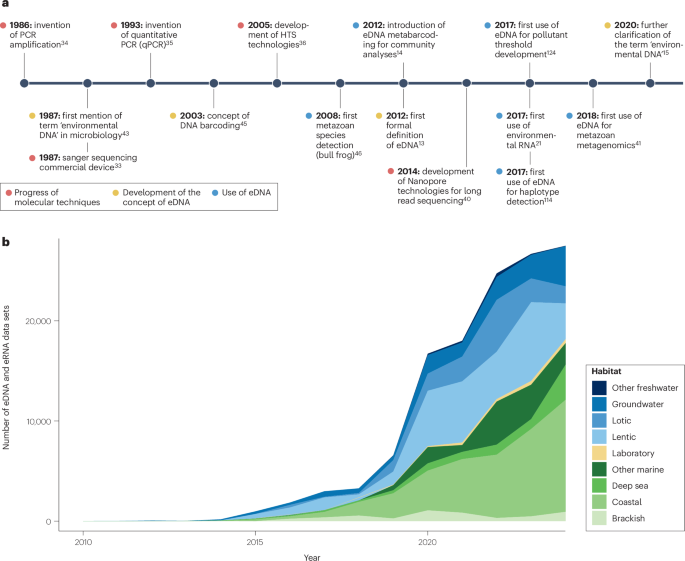
Following pioneering work in the 1980s and 1990s to isolate microbial communities from environmental samples43,44 and the development of DNA barcoding technology for species identification45, the combination of these technologies in the late 2000s46 led to eDNA becoming established as a research field in its own right13. Uptake of eDNA-based approaches has since vastly expanded in the field of biodiversity research owing to their non-invasiveness and increased efficiency for detecting species in a range of habitats at decreasing costs47. Technological advancements36,48 have enabled progression from detecting single species — initially focused on protected or invasive alien taxa46,49 — to comprehensive biodiversity assessments of many taxa with a metabarcoding approach. Consequently, the past 15 years have seen substantial growth in the number of datasets produced using this approach (Fig. 3), alongside the development of guidelines, large-scale projects and end-user applications15,29,37,47,50,51,52,53,54.
a, A schematic chronological timeline of important steps in the development of the concept of environmental DNA (eDNA) (yellow dots), progress of molecular techniques (red dots) and use of eDNA (blue dots). b, Temporal trends in aquatic data volume over time as the number of datasets published in the NCBI Sequence Read Archive on 20 September 2024. Here, a dataset is defined as a data package with a specific run number. Metadata on these datasets were retrieved using Entrez Programming Utilities and parsed using a list of keywords to include only eDNA and eRNA datasets from aquatic environments. HTS, high-throughput sequencing.
The uptake of eDNA research has been particularly prevalent in aquatic ecosystems, including rivers, lakes and marine water bodies, because of the ease of sample collection. Gaps in biodiversity knowledge are often larger for aquatic ecosystems than for terrestrial ecosystems, necessitating the implementation of novel approaches tailored to aquatic ecosystems. Current eDNA research in aquatic environments has mostly focused on freshwater ecosystems (65% of studies compared with 25% on marine ecosystems and 10% on other/multiple systems) and more than half of the studies have focused on fish (52%)51 — probably due to their high commercial and recreational values. Additionally, fish tend to shed greater amounts of DNA than species with sclerotized exoskeletons (such as crayfish) and are easier to detect owing to their high biomass and mobility55.
The primary strength of using eDNA analysis for assessing biodiversity is the ability to infer species’ presence without direct collection or observation13, and that the same approach can be applied across the whole tree of life. The minimally invasive sampling involved is desirable from ethical, work-safety and systems perspectives, as massive upscaling can be achieved without unwanted side effects. However, indirect inference — detecting DNA and not the organisms themselves — is sometimes also seen as the greatest weakness of eDNA analysis. This approach limits inference and information on population structure (abundance or life stages) and requires cautious interpretation of the signal as positive detections might be linked to transport of DNA56, sequencing errors57 or database classification errors58. Therefore, eDNA workflows require rigorous methods and best practices at each stage to minimize the risk of false positives59.
Using eDNA to support biodiversity policy
Successfully meeting the GBF targets requires the acquisition of three types of data (Fig. 1). First, biodiversity must be mapped across all scales and systems to identify priority areas for conservation and restoration (‘map biodiversity’; for example, GBF targets 3, 6 and 14). Second, the spatial movements and temporal changes in abundance of species of interest (such as pathogens, invasive alien or endangered species) must be tracked (‘track species’; for example, GBF targets 4 or 6). Finally, the responses of ecosystems to anthropogenic changes must be monitored and understood, including identifying pollution levels that do not harm biodiversity (as mentioned in GBF target 7) or tracking recovery success when restoring degraded ecosystems (see the ‘Understand and attribute drivers of biodiversity change’ section). The analysis of eDNA is a powerful approach to address all these aspects and to provide common information needed for implementing efficient regulation. In this section, the potential use of eDNA-based approaches for these three objectives is explored and examples of their usefulness for GBF targets presented.
Spatiotemporal mapping of aquatic biodiversity
Effective ecosystem assessment for policymaking, such as monitoring systems under pressure or identifying biodiversity hotspots, hinges upon efficient characterization of biodiversity, including adequate baseline data and mapping biodiversity trends over space and time60. Traditional sampling approaches such as gillnet fishing or trawling are still widely used, but are not sufficient to reach the ambitious targets of GBF and other regional initiatives (such as the European Union (EU) Water Framework Directive) because they are too labour intensive, destructive and costly to scale up or implement — particularly in undersampled regions, such as tropical streams, open ocean or groundwater systems. By contrast, eDNA analysis allows greater spatial resolution and increased temporal and taxonomical scales compared with traditional approaches, and eDNA sampling is increasingly adopted by public agencies as part of their routine biomonitoring programmes61,62,63. eDNA-based approaches effectively complement traditional aquatic biodiversity monitoring as they overcome the challenges of morphology-based classification (such as juvenile life stages or cryptic species64) and enable access to undersampled and less-accessible systems such as groundwater ecosystems65 or oceanic trenches66.
Technological advances are currently driving this transition. The ever-increasing pace of technological developments in eDNA science has inevitably reduced the cost of processing water samples67, enabling the collection of samples at high spatial and temporal resolution68,69. Although sample collection and eDNA capture can still be labour intensive, collecting water samples is becoming considerably faster owing to the development of autonomous robotic and passive samplers. Existing autonomous samplers70,71,72,73 are capable of multisample capture and preservation. When coupled with a microfluidic block, they can even collect, filter, extract and perform multiple targeted qPCR assays of eDNA samples in situ74,75. These automated samplers are highly attractive for early detection of invasive alien species (addressing GBF target 6; see the ‘Tracking endangered, harmful or invasive alien species’ section), and can be operated for months at a time without intervention76; however, further technological improvements are possible and the cost of fully automated samplers is currently prohibitive for some applications. Low-cost and less technologically advanced alternative samplers are based on passive capture of eDNA on substrates, such as non-charged cellulose ester or even medical gauze secured in three-dimensional hollow printed housing77,78. Traditionally, monitoring biodiversity was often restricted to specialists, whereas these simple eDNA capture methods have the potential to include citizen scientists in eDNA-based biodiversity assessment29,30,31, contributing to increased participation and improving integration of biodiversity into decision-making at every level (GBF targets 22 and 14, respectively).
Owing to transportation and mixing processes in water bodies, aquatic eDNA samples taken at a specific location reflect both the local community and organisms living in the surrounding environment. This feature can be leveraged to understand the spatial structure of biodiversity underpinning the observed data. In rivers, for example, depending on the discharge and other abiotic factors, eDNA can travel downstream from a few hundred metres to tens of kilometres56,79,80, acting as a ‘conveyor belt of biodiversity information’16. Thus, eDNA-based biodiversity measures not only correlate to upstream-averaged measures of land use and habitat types17,81,82,83, but also provide space-filling projections of biodiversity by integrating signals from discrete sampling points into a continuous representation along a watercourse (Fig. 2). Similarly, in marine systems, eDNA is transported by currents and shaped by the stratification of the water column84,85. Coupling stream86,87 and marine88,89 eDNA data with hydrodynamic models is therefore essential to unravel and understand the spatial distribution of biodiversity. Specific models90,91 enable spatial projection of pointwise eDNA samples beyond their collection point into space-filling taxon richness maps at high spatial resolution92. As such, they provide spatially integrated information on biodiversity.
The assessment of overall biodiversity is particularly relevant for GBF targets that focus on area-based or area-projected measures, such as target 1 (‘plan and manage all areas to reduce biodiversity loss’), target 3 (‘conserve 30% of land, water and seas’) or target 14 (‘integrate biodiversity into decision-making’). These targets are most effectively reached when overall biodiversity assessments of ponds, streams, groundwater and marine systems are comparable, implementable and accessible from local to global scales.
Tracking endangered, harmful or invasive alien species
Tracking or detecting species of particular interest or concern is often a key piece of information needed for policymaking and communication. In this context, eDNA analysis is a valuable tool for the detection and monitoring of individual species. Indeed, the first applications of eDNA were for the detection of either rare and endangered49 or invasive alien species46,93. The detection and monitoring of individual species of interest is directly associated with several GBF targets, including target 4 (‘halting extinction of [individual] endangered species’) and target 6 (‘reducing the introduction of invasive alien species’). Two examples of integrating eDNA information on targeted species into policy are the tracking of the endangered great crested newt in the UK using eDNA94 and the almost globally implemented eDNA (and eRNA) tracking of pathogens (in particular SARS-CoV-2) in wastewater samples95. In both cases, real-time implementation and analysis of eDNA samples enables rapid dissemination to inform policy. eDNA-based methods are now implemented in many countries for routine monitoring of harmful algal blooms96,97, invasive alien species, such as the quagga mussel98 and European green crab99, and of endangered species such as the above-mentioned great crested newt94 or the harbour porpoise49.
Importantly, however, an eDNA signal does not automatically indicate an organism’s immediate temporal or spatial occurrence. The signal can be transported in space (for example, by water current or in the faeces of predators), reflect past occurrences (ancient DNA), be a transient signal of an organism passing by, or even enter the water via wastewater or terrestrial run-off16,100,101,102. An eDNA signal therefore contains different information compared with the direct observation of an organism, including potential false positives103,104, and thus requires different interpretation. For example, detection of an alien species using eDNA analysis in a location where it has not previously been observed should not trigger a policy response for removal, but rather incentivize the deployment of other methods to confirm the species’ presence. Similarly, eDNA-based techniques do not allow the collection of population demographics, such as age and size distribution or sex ratio, which can be required for endangered or invasive alien species monitoring105. Some of these limitations can be overcome by sampling eRNA rather than eDNA, as eRNA represents genes expressed in the sampled environment and degrades faster than eDNA20. As eRNA provides a more contemporary signal than eDNA, its analysis can potentially distinguish between live and dead material to reduce the number of false positives18,21 or, in specific cases, even differentiate life stages (for example, in amphibians). However, eRNA analysis is still under evaluation and, with a few exceptions19, is not yet ready for use in monitoring contexts to inform policymakers.
Approaches based on eDNA are potentially relevant for GBF target 4, which specifically links the halt of species extinctions to the prevention of genetic diversity loss. Addressing this target currently relies on indirect measures of genetic diversity based on abundance data for many species and subsequent estimation of effective population size. Determining abundance estimates from eDNA is challenging, but at least possible in some circumstances. Both targeted106,107 and metabarcoding108 approaches have demonstrated correlation between copy number and/or read counts from eDNA and biomass or abundance, indicating that the method is at least semi-quantitative. However, this approach is strongly debated owing to the high potential for bias throughout eDNA workflows, which can create noise and error in the signal109,110,111,112,113. Possible solutions include adding internal standards of a known amount of synthetic DNA (so-called DNA spike-in) to compare read counts within species and across sites.
In addition to indirect measures of genetic diversity based on abundance, eDNA analysis could enable direct measurement of intraspecific genetic diversity, even in hard-to-sample environments and for rare and elusive species. This approach is still in development and is not yet ready for direct implementation; however, it has shown promising results in particular cases. So far, investigations have focused on the detection of haplotypes in short sections of mitochondrial (mt)DNA used for metabarcoding, and demonstrated that eDNA captures the genetic diversity of sampled populations38,114,115,116,117. Such approaches can differentiate populations and identify regions with unique genetic variation based on the mtDNA marker genes118. However, these short mtDNA fragments generally have low power to resolve finer genetic structures within or between populations. Targeting longer fragments and nuclear DNA provides increased resolution but presents additional challenges related to the shorter persistence of longer DNA fragments in the environment, and that eDNA represents a mixture of DNA from different individuals. Current methods do not allow assigning genotypes to individuals, which restricts most genetic approaches to population-level estimates119. Likelihood-based DNA mixture models could be applied to address the problem of redundancy (multiple individuals sharing the same allele) as long as a relatively large panel of multi-allelic markers is available120. Technical challenges notwithstanding, microsatellite allele frequencies of round gobies (Neogobius melanostomus) or an Arctic diatom (Thalassiosira hyalina) estimated from aquatic eDNA have been shown to closely resemble allele frequencies from genotyped tissues120,121.
A considerable challenge for estimating genetic diversity from eDNA data is the detection of errors generated during the amplification and sequencing steps. For species detection, many bioinformatic procedures remove genetic variability during regular data cleansing and cluster sequences at the species level. For population genetic-level analysis, keeping exact sequence variants while removing erroneous sequences is the primary goal and can be more challenging119. Resolving this challenge requires combined improvements in sequencing technologies and the computational steps to identify and remove errors.
Understand and attribute drivers of biodiversity change
For policymaking and management, attribution of biodiversity change is key: only when drivers of change (both positive and negative) are linked to their biodiversity effects can effective policy and management be achieved. Traditionally, attributing drivers to aquatic biodiversity change has relied on monitoring and morphological identification of a relatively small number of species (such as macroinvertebrates, benthic macrofauna, fish and diatoms) that are biological indicators of environmental pressures such as nutrient pollution122. However, these methods are often unsuitable for monitoring emerging drivers, such as pollution by nanoparticles or microplastics, and hinge on the (often incorrect) assumption that a few indicator groups represent overall ecosystem responses. eDNA-based methods have already led to marked improvements in our ability to document biodiversity123 and identify pollutant thresholds124 by integrating data from diverse taxonomic groups with only one sample. This approach, in comparison with traditional methods, will provide a more comprehensive — and at least more diversified — response of aquatic communities to anthropogenic changes, individually and in combination24,25,125.
Current monitoring, whether with traditional or molecular tools, often focuses on the structure of communities (their richness and composition) rather than their functional properties123. However, the effect of biodiversity loss on ecosystem services cannot be fully understood from estimates of taxonomic diversity alone. To characterize ecosystem health in its entirety, structural and functional biodiversity must be measured as these parameters can respond differently to different stressors. This type of monitoring requires a shift from focusing on which species provides the function to understanding how the functional properties of the whole community respond to pressures123,126. For example, co-occurrence data generated from eDNA metabarcoding across the tree of life allow reconstructing multitrophic ecological networks, from which functional properties such as connectance and nestedness can be estimated127. Other emerging approaches for addressing ecosystem changes are based on the analysis of expressed genes via eRNA. The strength of these eRNA methods lies in monitoring the physiological state of populations or communities by detecting changes in the activity of single or multiple genes18. Shotgun sequencing of total RNA from environmental samples (metatranscriptomics) allows the identification of genes that are actively and collectively expressed under different conditions128. Changes in freshwater biofilm microbial community-level expression in response to pollutants have been demonstrated, even when changes in community composition were not detected129. Additionally, experiments have revealed changes in community-wide gene expression of aquatic organisms under heat stress through sampling experimental tank water, demonstrating that metatranscriptomics of extra-organismal eRNA can reveal gene expression response to environmental change130. These emerging measures of ecological condition, especially when using eRNA, are promising but not yet ready for widespread use for practitioners and to inform decision-making.
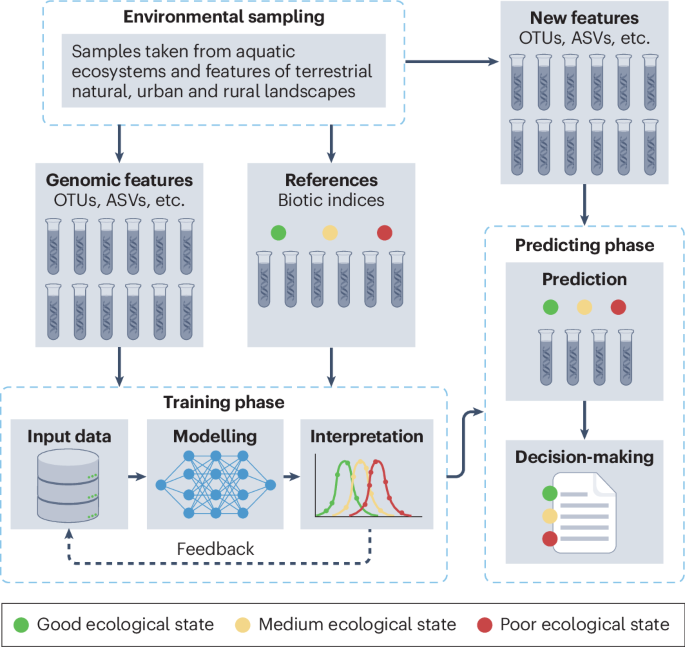
Advances in artificial intelligence and modelling are enabling new approaches for attributing biodiversity change to environmental and/or anthropogenic drivers. For example, eDNA coupled with machine-learning models is used to infer the ecological status of aquatic systems131,132, and allows inclusion of a broader set of indicator taxa. Microorganisms, for example, differ considerably in their metabolism and sensitivity to different pollutant sources133, but have been largely ignored in biomonitoring because they are difficult to culture and identify, and because knowledge of their ecological function is unavailable134,135. Validated approaches based on profiling communities using amplicon sequence variants and/or operational taxonomic units detect changes in the structure of communities of bacteria133,136, freshwater invertebrates25,137 and diatoms138 against well-understood disturbance gradients, and have demonstrated strong improvements in the accuracy of ecological quality assessment139,140. Supervised machine learning combined with metabarcoding will complement and expand current morphology-based methods to produce the same ecological quality assessments at increased speed and cost efficiency, and without necessarily depending on taxonomic assignment using barcode reference databases135 (Fig. 4). The ability of supervised machine learning to outperform traditional indicator value approaches, using training datasets with reference disturbance levels to estimate biotic indices, has been demonstrated131,132,140,141,142,143.
The principle of the machine-learning approach is to link genomic features of the environmental DNA (eDNA) samples (such as operational taxonomic units (OTUs), amplicon sequence variants (ASVs) or taxonomically assigned information) to environmental reference states. Specifically, during the training phase, the model learns relationships between genomic features and known environmental conditions, iteratively refining its predictive rules. In the predicting phase, the trained model applies these learned rules to analyse new eDNA samples, generating straightforward, actionable predictions. Through continuous feedback cycles, the model enhances its accuracy, improving its utility for environmental monitoring and management.
Overall, eDNA analysis enables attributing changes in biodiversity to specific environmental factors17,65,144, evaluating the impact of implementing protected areas145,146, and calculating biodiversity and ecosystem state indices47. Aquatic eDNA analysis can also attribute change beyond aquatic ecosystems: imprints of terrestrial land use on aquatic eDNA are well established, showing that aquatic eDNA signals associate to the structure and diversity of terrestrial land use assessed by remote sensing at 400–1,000-m upstream distances17. This information can then be translated into effective practice for the prevention of biodiversity decline.
Examples of eDNA use for policy and beyond
eDNA-based biodiversity monitoring has the potential to scale from a local to a regional and eventually a global perspective, and enables data integration and the use of this information to downscale back to regional and local interventions42. eDNA approaches are already widely used across different scales, which helps to contribute to monitoring progress on GBF targets. These methods are particularly useful for spearheading activities in regions in which previous knowledge on biodiversity is limited; for example, eDNA-based approaches are being tested to study the biodiversity of deep marine habitats147,148. Similarly, subterranean habitats are one of the least studied environments and the use of molecular tools such as eDNA contributes to mapping their biodiversity65,149. Moreover, eDNA-based assessments are being developed and implemented in many of the world’s largest rivers such as the Danube150, Yangtze144 or Rhine151, which are not only home to enigmatic megafauna but are also severely threatened by anthropogenic activities.
Achieving target 1 of the GBF (‘plan and manage all areas to reduce biodiversity loss’) depends on understanding the initial state and change of overall biodiversity. In many cases, eDNA data are the very first baseline data, and linking these data to possible drivers of biodiversity change will ultimately guide restoration planning152. The ability of eDNA analysis to provide data on the state and change of individual species focuses on target 4 (‘halt species extinction, protect genetic diversity, and manage human–wildlife conflicts’), which directly refers and links to the monitoring and managing of biodiversity, at the species or within-species level. Similarly, target 6 (‘reduce the introduction of invasive alien species by 50% and minimise their impact’) operates at the species level, aiming to decrease the impact of alien species in part through preventing their introduction in aquatic systems. To support this approach, eDNA analysis provides early detection and monitoring of alien species. Although initial attempts to detect invasive alien species (for example, the Asian carps Hypophthalmichthys nobilis and Hypophthalmichthys molitrix93,153) were partially challenged by detection failures (false absences), false presence indications and unknown lower thresholds of detectability, increasing standardization and the adoption of structured decision-making processes have resulted in a relatively broad application of eDNA methods to monitor Asian carp species in North America23,154. Similar efforts have been established for other aquatic alien species, such as invasive kelp (Undaria pinnatifida) and sea stars (Asterias amurensis)155, tunicates (Didemnum vexillum)156, dreissenid mussels (Dreissena spp.)22 or freshwater aquatic plants (Najas minor)157.
The contribution of eDNA analysis to GBF targets 1, 4 and 6 focuses on simply detecting the presence of species. However, eDNA analysis can provide insight into why and how biodiversity is affected; that is, mechanistic monitoring of aquatic systems and the effects of stressors. Classic assessments of environmental toxicology and environmental chemistry have focused on monitoring a highly diversified set of chemicals10. To reach target 7 (‘reduce pollution to levels that are not harmful to biodiversity’), pollutant monitoring must be linked to an ecosystem’s ecological health and resilience status. eDNA is effective in capturing how environmental stressors, particularly chemical pollution, have combined negative effects on the structure, stability and composition of aquatic communities across a wide range of taxa158,159,160. Target 10 (‘enhance biodiversity and sustainability in agriculture, aquaculture, fisheries, and forestry’) requires similar monitoring of effects on biodiversity, but in relation to the production and harvesting of natural resources. eDNA is already routinely used to monitor impacts of aquaculture (such as salmon farming) by examining marine benthic sediments161 and has also been used to monitor the outflow of wastewater into natural streams, focusing on possible effects of different agricultural runoff100.
Next steps for eDNA in the GBF context
eDNA-based methods have the potential to greatly improve our understanding of aquatic ecosystems and contribute to national and global biodiversity targets. For successful inclusion of eDNA analysis in monitoring programmes, careful action is needed as methods must be broadly applicable, comparable and scalable, and must be implementable for countries across the globe.
Two key barriers exist to the implementation of eDNA: first, the perceived limited comparability between results generated with eDNA-based approaches and those obtained with traditional methods162,163,164; and second, unequal access to technical infrastructure. eDNA metabarcoding technology is considered ready for implementation and allows — when needed — retrofitting of indices to current standards47,135. Discrepancies with traditional assessments occur because of amplification biases and incomplete reference databases, but also because eDNA can detect cryptic or elusive species47,123,165 previously missed by traditional tools. Assessing biodiversity and achieving successful conservation outcomes will require effectively using eDNA alongside traditional methods, as biodiversity monitoring globally remains under-resourced. In high-income countries, existing monitoring programmes should be complemented with eDNA-based approaches, creating continuity for existing long-term monitoring and generally allowing comparison at the index level. Maintenance and overlap with other programmes will increase costs and require extra work over the short term, but secure existing data series in the long term. To encourage the rapid adoption of eDNA globally, in low- and middle-income countries, novel (and first) monitoring could use eDNA only.
This section discusses necessary steps towards a global application of eDNA to assess biodiversity within the context of the GBF. First, methods of standardization must be agreed and promoted (agree on minimal standards on sampling and processing, provide training opportunities and institutionalize quality control). Second, adequate reference databases, digital sequence data deposition and general access to these resources must be established. Finally, efforts must be made to increase inclusivity and remove technology barriers (collective benefit, authority to control, responsibility and ethics (CARE) principles and Nagoya Protocol).
Standardization
The development and implementation of international standards is an essential next step to make eDNA approaches broadly applicable. These next steps, including establishing standardized protocols and best practices for general implementation of eDNA, are particularly relevant for global initiatives, such as the GBF, that rely on common agreement beyond the borders and jurisdiction of individual countries.
eDNA has always been developed as a tool for practitioners, rather than for academic research only. Consequently, discussions on an adequate and replicable implementation paralleled the rapid advances in the field153,162,166,167. General protocols and guidelines were developed from the first species-based assessments, such as amphibians in the UK29, to the monitoring of whole communities of invertebrates and fish, for example, in Switzerland168, the EU37,169, China170 or the USA93.
Despite these standardization efforts, eDNA-based biodiversity assessments still use various approaches throughout the world171. The selection of a certain filter material, DNA extraction methods, primers, sequencing technologies and post-processing steps can drastically affect the inferred taxa lists172,173. For multispecies detection approaches, such as eDNA metabarcoding, the effects of the choice of methods are most pronounced for rare species that are easily missed. Broader use of eDNA for monitoring thus requires standardization of practices63,174, in particular a global agreement on general minimum standards for sampling and processing, reporting and data publishing. Here, specific recommendations for different freshwater ecosystems (for example, small stream versus large, stratified lake), environmental factors (turbidity, flow, pH and so on) and specific research questions and tasks (single species detection versus multispecies detection) must be accounted for37,168,175. This standardization is well underway — a first ISO standard is available in draft form after international voting176, and several others are in development under the framework ISO/TC 147/SC 5 (Biological methods) — but must be continued. Given the ambitious 2030 GBF timeline, the implementation and standardization of the method in its current form might be more effective than deferring implementation considering the promise of upcoming new technologies.
Populating reference databases, FAIR curation of data and scalability
Comparability and scalability across taxa and regions are the biggest assets of eDNA analysis. These capabilities hinge, however, on adequate access to reference databases as most uses of eDNA rely on assigning specific sequences to taxonomic and genetic reference databases (although taxonomy-free approaches exist139). These reference databases are globally still highly unevenly distributed, and large geographic and taxonomic gaps are present177,178,179. For example, (sub)tropical systems and many invertebrate groups are far less represented than temperate systems and vertebrate groups. Populating, expanding and maintaining these reference databases is imperative.
Advances in sequencing technologies enable the upscaling of reference database generation by several orders of magnitude while also drastically reducing costs. However, many — even formally described — species are still absent from public genomic repositories180. Novel nanopore sequencing technologies could allow the barcoding of thousands of specimens from a single amplicon pool at a sequencing cost of few US$ per specimen, offering the potential to catalogue all species within two decades181. The portability and applicability of sequencing technologies in field conditions is also rapidly improving; for example, portable sequencers have been used for DNA barcoding in the Ecuadorian Chocó Rainforest182 and other remote locations. Populating databases is still a major bottleneck as implementation is lagging behind this progress in technologies, and also critically depends on taxonomists’ knowledge and expertise. Scientists and end-users must be mobilized alike and commit to complementing the databases as the task is important, necessary and cannot be delegated. This challenge is particularly pronounced given the large number of undescribed or underrepresented taxa, discrepancies in taxonomic classifications and the need for rigorous validation of reference sequences. The complexity of maintaining and curating these databases has been widely discussed in the literature, touching on issues such as funding177 and standardization efforts58. A comprehensive discussion of these aspects is, however, beyond the scope of this Perspective.
Alongside data generation, the raw data produced from environmental samples must be deposited in repositories (Table 1) whose long-term operability is guaranteed183. Priority should be given to open platforms specifically adapted to host genetic data, enabling better discoverability through advanced application programming interfaces (which enable the exchange of data between existing programmes) and research tools184. The International Nucleotide Sequence Database Collaboration, linking the Sequence Read Archive, the European Nucleotide Archive and the DDBJ Sequence Read Archive are currently among the most suitable options for depositing and indexing eDNA data. Ideally, eDNA repositories should facilitate the seamless exchange and integration of data across different platforms and disciplines. For example, biodiversity data portals such as the Ocean Biodiversity Information System (OBIS) and the Global Biodiversity Information Facility (GBIF) can include biodiversity records derived from raw DNA data while providing a permanent link to the original repositories185. Publication of eDNA datasets in these global biodiversity databases is crucial to ensure that data can be used by policymakers with little to no specialist knowledge186. However, so far, few eDNA datasets have been made publicly available through these platforms. The key to successful adoption is the mandatory deposition of eDNA raw data along with relevant metadata provided in standardized data models, such as provided by the Genomics Standards Consortium (see the MIxS MIMARKS checklists187) or the Biodiversity Information Standards consortium TDWG (Darwin Core extended data model188,189). As part of ongoing international task forces, new metadata standards are currently being developed190. These data standards ensure that the most critical metadata are recorded and made available through a standardized vocabulary191, which in turn can be aligned with other biodiversity community standards. Such metadata (Table 1) are crucial as they ensure that the data can be accurately interpreted and reused in various contexts, which is necessary to reach a broad range of stakeholders, including citizens.
eDNA data producers must focus on creating datasets that can be used and reused186, which involves ensuring the longevity of data, adopting standard formats and metadata for ease of interpretation, and promoting open access to facilitate wide usage across different sectors. Thus, published eDNA data must adhere to the FAIR principles — findable, accessible, interoperable and reusable192 — as this approach enhances the discoverability and usability of eDNA data across various scientific and non-scientific domains186 (Table 1). Therefore, setting minimum guidelines for data reporting and analysis is recommended, and minimum information to be reported as part of any aquatic eDNA study has been proposed190,193. Data deposition must include the raw sequencing data and the essential metadata specific to the samples and their processing in the laboratory, but also the final community data matrix, easily interpretable by the stakeholders, as well as the bioinformatics tools and reference databases required to produce the results. These minimum guidelines will ensure a baseline of quality and comparability for eDNA research and application.
CARE and inclusivity
The successful, widespread implementation of eDNA hinges on removing barriers to its access and making the methods inclusive. Technological barriers and uneven distribution of knowledge and resources between high-income countries and low- and middle-income countries exist across biodiversity sciences. As eDNA analysis (similar to remote sensing and other technology-driven approaches) depends on specialized laboratory and analysis techniques, overcoming discrepancies in lack of knowledge, training and resources is particularly relevant. If countries cannot afford eDNA-based monitoring or do not have access to the technologies to process and assess the samples, the existing inequality in biodiversity knowledge will further increase.
In addition to ensuring equitable access to eDNA technology, local and Indigenous knowledge on biodiversity must be both valued and integrated into technology-driven monitoring efforts. Although measuring biodiversity variables through proxies such as eDNA is technologically feasible, maintaining taxonomic and natural history knowledge, as well as connections to ecosystems and nature, is essential. Datafication of biodiversity and reliance on technological solutions run the risk of disconnecting people with nature194, as sequencing information and eDNA approaches cannot replace observing and experiencing nature directly. Additionally, limits exist to the quality and applicability of data available from methods based on indirect collection of information, such as remote sensing or eDNA. Conversely, eDNA also has the potential to be a powerful tool that provides people with a new perspective on their surrounding environment and biodiversity, strengthening their interest in and commitment to nature29,30,31. Any eDNA-based approaches should, therefore, consider how to include the public and taxonomic experts such that the information on biodiversity from eDNA is accompanied by increased understanding and perception of biodiversity.
Addressing the sociocultural dimensions of eDNA, particularly when data originate from territories of Indigenous communities, necessitates the adoption of the CARE principles195. These principles ensure that eDNA research is conducted in a manner that respects the rights and traditions of local communities, promoting an ethical approach and facilitating the local populations acceptance of biodiversity research. Incorporating the CARE principles aligns eDNA practices with broader societal values and legal frameworks, such as the Nagoya Protocol196 and GBF target 13 (‘fair and equitable sharing of genetic resources’). In some situations, however, open-access goals such as the population and maintenance of reference databases can bypass traditional mechanisms for benefit sharing, potentially disadvantaging the countries or communities where the genetic material originated. Public deposition without consultation can undermine the authority of these communities over resources tied to their cultural identity and governance systems. Moreover, eDNA can also complicate traditional frameworks, especially when DNA is transported across national boundaries by natural processes such as ocean currents, river flows and air197. All stakeholders — nations, Indigenous groups and local communities — must be included in discussions about the implementation of CARE principles, data sharing and benefit sharing to ensure that the benefits of eDNA research are distributed fairly and with due regard to the sovereignty and cultural significance of the data sources194,198.
Biodiversity credits and the marketing of biodiversity
One of the four overarching goals of the GBF is to mobilize the business and finance sectors to close the gap in funding biodiversity. In particular, target 19 (‘Mobilize $200 billion per year for biodiversity’) encourages solutions to incentivize international companies and finance to engage in the monitoring of biodiversity and contribute monetarily to its protection. Several initiatives are being promoted to market biodiversity and to incentivize the development of activities and technologies that are not harmful for the environment or that mitigate deleterious actions, such as green bonds or biodiversity credits (quantifiable units of biodiversity improvement that should provide an incentive for funding conservation projects)199,200. Although such market-based solutions to environmental challenges are gaining substantial attention at the global scale, deep uncertainties exist regarding the process and their benefits199. Without proper transparency and accountability, these schemes might be ineffective and lead to greenwashing and inequitable outcomes, as has occurred for carbon credits201.
A challenge to the broad use of biodiversity credits is the difficulty in quantify biodiversity improvements accurately and in a standardized manner. Sampling and identification methods vary substantially, are not all easily implementable across the globe, and the multidimensionality of biodiversity cannot be captured in one single number. eDNA metabarcoding has been suggested as a useful method to help the development of marketing metrics for biodiversity, as its simplicity and scalability could allow the collection of comparable data across the globe. However, this approach also requires global standardization of methods and a clear reference to its limits. Moreover, eDNA is particularly well suited for aquatic environments but is less proven for terrestrial ecosystems, which might limit the comparability of this data201,202.
Summary and future directions
Addressing biodiversity decline and implementing global measures to assess, manage and restore biodiversity are essential and at the core of the GBF framework. The huge progress in molecular-based biodiversity monitoring, particularly the use of eDNA, has created the first feasible and coherent approach to monitor aquatic biodiversity. To successfully implement eDNA biomonitoring as a reliable and widely trusted basis for policy and decision-making, a new common agreement on its use is needed, including common protocols, data standards and standardization for quality assurance. Further, implementation of eDNA approaches hinges on increased FAIRness and open accessibility of biodiversity data globally, which also presents an opportunity to enhance collaboration, improve conservation efforts and advance research. Rapid completion of reference database, curation and integration of FAIR principles are needed. Although progress towards these goals, in particular where and by whom data are stored, is becoming more systematically addressed in academic research, agreement has not yet been reached and implemented regarding data produced by public stakeholders and private companies. Sample repositories (such as the Global Genome Biodiversity Network) and data centres established for eDNA data should not only be accessible and implemented for high-income countries (for example, the International Nucleotide Sequence Database Collaboration), but also for low- and middle-income countries. As these repositories are funded by public money and provide collective benefits, all data collected through governmental programmes should be included — even when data collection and analysis is executed by private companies.
Coherent, integrated and complete data storage and access will allow establishing new universally valid metrics for describing biodiversity and elevate the quality and comparability of biodiversity data. In the framework of the GBF, regional to global progress on reaching targets could be tracked by new eDNA-based indicators on the state, change and pressures of biodiversity. Particularly in aquatic systems, these metrics can be calculated at high spatial and temporal resolutions: transport of eDNA in the water allows predictions of biodiversity across catchments and water bodies, whereas matching eDNA sampling to existing monitoring stations of water quality allows synergy with existing infrastructure. A pragmatic approach should be chosen when designing new metrics versus retrofitting to existing monitoring programmes: that is, complementing rather than replacing existing monitoring programmes, yet building new monitoring timelines where no data are available. Swiftly establishing a global network of coherent aquatic biodiversity monitoring using eDNA is realistic and required to achieve GBF targets.
A possible way forwards is institutionalizing eDNA monitoring activities through regional centres (examples of such attempts include the Defra DNA Center of Excellence in the UK, the Joint Research Center in the EU and the National eDNA Reference Centre in Australia). These centres coordinate activities that include funding, supporting and driving research that is relevant to policy implementation, reaching out to local monitoring personnel, receiving samples from citizens, performing quality checks through internal positive control, and generating reference material and certification standards. Such regional eDNA excellence centres can facilitate capacity building and act as coordinating and executing building blocks, linking both local and global biodiversity monitoring centres (similar to GEO BON). The Canadian Centre of Biodiversity Genomics at the University of Guelph and other federal initiatives are examples of such centres. Global join up of these initiatives and centres is crucial to ensure international standards and comparability of datasets across countries. Finally, to future proof the data acquired and ensure long-term comparability, adequate biobanking of samples is a necessary next step, which should, at a minimum, include storing metadata, raw sequencing output, and the extracted DNA samples in quality and quantity sufficient to be used by generations to come203. Together, these measures will elevate the accessibility and extent of aquatic biodiversity data to levels needed for achieving GBF targets and securing global biodiversity.
References
-
Keck, F. et al. The global human impact on biodiversity. Nature https://doi.org/10.1038/s41586-025-08752-2 (2025).
-
Pereira, H. M. et al. Global trends and scenarios for terrestrial biodiversity and ecosystem services from 1900 to 2050. Science 384, 458–465 (2024).
-
Loreau, M. et al. Do not downplay biodiversity loss. Nature 601, E27–E28 (2022).
-
Rockström, J. et al. A safe operating space for humanity. Nature 461, 472–475 (2009).
-
World Economic Forum. Global Risks Report 2022, 17th edn (WEF, 2022).
-
United Nations Convention on Biological Diversity. Kunming–Montreal Global Biodiversity Framework. CBD https://www.cbd.int/doc/decisions/cop-15/cop-15-dec-04-en.pdf (UN, 2022).
-
Gonzalez, A. et al. A global biodiversity observing system to unite monitoring and guide action. Nat. Ecol. Evol. 7, 1947–1952 (2023).
-
Almond, R. E., Grooten, M. & Peterson, T. World Wildlife Fund. Living Planet Report 2020 — Bending the Curve of Biodiversity Loss (WWF, 2020).
-
Intergovernmental Science–Policy Platform on Biodiversity and Ecosystem Services. Global assessment report on biodiversity and ecosystem services (IPBES, 2019).
-
Schwarzenbach, R. P. et al. The challenge of micropollutants in aquatic systems. Science 313, 1072–1077 (2006).
-
Vörösmarty, C. J. et al. Global threats to human water security and river biodiversity. Nature 467, 555–561 (2010).
-
Leclère, D. et al. Bending the curve of terrestrial biodiversity needs an integrated strategy. Nature 585, 551–556 (2020).
-
Taberlet, P., Coissac, E., Hajibabaei, M. & Rieseberg, L. H. Environmental DNA. Mol. Ecol. 21, 1789–1793 (2012).
-
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C. & Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050 (2012).
-
Pawlowski, J., Apothéloz-Perret-Gentil, L. & Altermatt, F. Environmental DNA: what’s behind the term? Clarifying the terminology and recommendations for its future use in biomonitoring. Mol. Ecol. 29, 4258–4264 (2020).
-
Deiner, K., Fronhofer, E. A., Mächler, E., Walser, J.-C. & Altermatt, F. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat. Commun. 7, 12544 (2016).
-
Zhang, H. et al. A spatial fingerprint of land–water linkage of biodiversity uncovered by remote sensing and environmental DNA. Sci. Total Environ. 867, 161365 (2023).
-
Yates, M. C., Derry, A. M. & Cristescu, M. E. Environmental RNA: a revolution in ecological resolution? Trends Ecol. Evol. 36, 601–609 (2021).
-
Visco, J. A. et al. Environmental monitoring: inferring the diatom index from next-generation sequencing data. Environ. Sci. Technol. 49, 7597–7605 (2015).
-
Kagzi, K., Hechler, R. M., Fussmann, G. F. & Cristescu, M. E. Environmental RNA degrades more rapidly than environmental DNA across a broad range of pH conditions. Mol. Ecol. Resour. 22, 2640–2650 (2022).
-
Pochon, X., Zaiko, A., Fletcher, L. M., Laroche, O. & Wood, S. A. Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLoS ONE 12, e0187636 (2017).
-
Sepulveda, A. et al. Using structured decision making to evaluate potential management responses to detection of dreissenid mussel (Dreissena spp.) environmental DNA. Manag. Biol. Invasion 13, 344–368 (2022).
-
US Fish and Wildlife Service. Great Lakes eDNA monitoring program. Asian carp Canada https://www.asiancarp.ca/surveillance-prevention-and-response/great-lakes-edna-monitoring-program/ (US FWS, 2020).
-
Romero, F., Acuña, V. & Sabater, S. Multiple stressors determine community structure and estimated function of river biofilm bacteria. Appl. Environ. Microbiol. 86, e00291–e00320 (2020).
-
Beermann, A. J., Zizka, V. M. A., Elbrecht, V., Baranov, V. & Leese, F. DNA metabarcoding reveals the complex and hidden responses of chironomids to multiple stressors. Environ. Sci. Eur. 30, 26 (2018).
-
Fediajevaite, J., Priestley, V., Arnold, R. & Savolainen, V. Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecol. Evol. 11, 4803–4815 (2021).
-
Buchner, D., Macher, T.-H., Beermann, A. J., Werner, M.-T. & Leese, F. Standardized high-throughput biomonitoring using DNA metabarcoding: strategies for the adoption of automated liquid handlers. Environ. Sci. Ecotechnol. 8, 100122 (2021).
-
Ruppert, K. M., Kline, R. J. & Rahman, M. S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547 (2019).
-
Biggs, J. et al. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv. 183, 19–28 (2015).
-
Larson, E. R. et al. From eDNA to citizen science: emerging tools for the early detection of invasive species. Front. Ecol. Environ. 18, 194–202 (2020).
-
Couton, M. et al. Integrating citizen science and environmental DNA metabarcoding to study biodiversity of groundwater amphipods in Switzerland. Sci. Rep. 13, 18097 (2023).
-
Deiner, K. et al. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895 (2017).
-
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA 74, 5463–5467 (1977).
-
Mullis, K. et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 51, 263–273 (1986).
-
Higuchi, R., Fockler, C., Dollinger, G. & Watson, R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology 11, 1026–1030 (1993).
-
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
-
Bruce, K. et al. A Practical Guide to DNA-Based Methods for Biodiversity Assessment (Pensoft, 2021).
-
Sigsgaard, E. E. et al. Population-level inferences from environmental DNA — current status and future perspectives. Evol. Appl. 13, 245–262 (2020).
-
Abad-Recio, I. L., Alonso-Sáez, L. & Lanzén, A. Toward functional profiling for eDNA�based monitoring in coastal environments: a comparison of three approaches. Environ. DNA 6, e504 (2024).
-
MacKenzie, M. & Argyropoulos, C. An introduction to nanopore sequencing: past, present, and future considerations. Micromachines 14, 459 (2023).
-
Bovo, S. et al. Shotgun metagenomics of honey DNA: evaluation of a methodological approach to describe a multi-kingdom honey bee derived environmental DNA signature. PLoS ONE 13, e0205575 (2018).
-
Thomsen, P. F., Jensen, M. R. & Sigsgaard, E. E. A vision for global eDNA-based monitoring in a changing world. Cell 187, 4444–4448 (2024).
-
Ogram, A., Sayler, G. S. & Barkay, T. The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 7, 57–66 (1987).
-
Steffan, R. J., Goksøyr, J., Bej, A. K. & Atlas, R. M. Recovery of DNA from soils and sediments. Appl. Environ. Microbiol. 54, 2908–2915 (1988).
-
Hebert, P. D. N., Cywinska, A., Ball, S. L. & deWaard, J. R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321 (2003).
-
Ficetola, G. F., Miaud, C., Pompanon, F. & Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425 (2008).
-
Blackman, R. et al. Environmental DNA: the next chapter. Mol. Ecol. 33, e17355 (2024).
-
Satam, H. et al. Next-generation sequencing technology: current trends and advancements. Biology 2023, 997 (2023).
-
Foote, A. D. et al. Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 7, e41781 (2012).
-
Leese, F. et al. DNAqua-Net: developing new genetic tools for bioassessment and monitoring of aquatic ecosystems in Europe. Res. Ideas Outcomes 2, e11321 (2016).
-
Takahashi, M. et al. Aquatic environmental DNA: a review of the macro-organismal biomonitoring revolution. Sci. Total Environ. 873, 162322 (2023).
-
De Brauwer, M. et al. Best practice guidelines for environmental DNA biomonitoring in Australia and New Zealand. Environ. DNA 5, 417–423 (2023).
-
Ferrante, J. et al. Gaining decision-maker confidence through community consensus: developing environmental DNA standards for data display on the USGS Nonindigenous Aquatic Species database. Manag. Biol. Invasion 13, 809–832 (2022).
-
Minamoto, T. et al. An illustrated manual for environmental DNA research: water sampling guidelines and experimental protocols. Environ. DNA 3, 8–13 (2021).
-
Andruszkiewicz Allan, E., Zhang, W. G., C. Lavery, A. & Govindarajan, F. A. Environmental DNA shedding and decay rates from diverse animal forms and thermal regimes. Environ. DNA 3, 492–514 (2021).
-
Deiner, K. & Altermatt, F. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9, e88786 (2014).
-
Burian, A. et al. Improving the reliability of eDNA data interpretation. Mol. Ecol. Resour. 21, 1422–1433 (2021).
-
Keck, F., Couton, M. & Altermatt, F. Navigating the seven challenges of taxonomic reference databases in metabarcoding analyses. Mol. Ecol. Resour. https://doi.org/10.1111/1755-0998.13746 (2022).
-
Goldberg, C. S., Strickler, K. M. & Pilliod, D. S. Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol. Conserv. 183, 1–3 (2015).
-
Gonzalez, A. & Londoño, M. C. Monitor biodiversity for action. Science 378, 1147 (2022).
-
Norros, V. et al. Roadmap for implementing environmental DNA (eDNA) and other molecular monitoring methods in Finland — vision and action plan for 2022–2025 (Finnish Environment Institute, 2022).
-
Blancher, P. et al. A strategy for successful integration of DNA-based methods in aquatic monitoring. MBMG 6, e85652 (2022).
-
Kelly, R. P. et al. Toward a national eDNA strategy for the United States. Environ. DNA https://doi.org/10.1002/edn3.432 (2024).
-
Mason, D. H. et al. Certain detection of uncertain taxa: eDNA detection of a cryptic mountain sucker (Pantosteus jordani) in the Upper Missouri River, USA. Environ. DNA 3, 449–457 (2021).
-
Couton, M., Hürlemann, S., Studer, A., Alther, R. & Altermatt, F. Groundwater environmental DNA metabarcoding reveals hidden diversity and reflects land-use and geology. Mol. Ecol. 32, 3497–3512 (2023).
-
Laroche, O., Kersten, O., Smith, C. R. & Goetze, E. From sea surface to seafloor: a benthic allochthonous eDNA survey for the abyssal ocean. Front. Mar. Sci. https://doi.org/10.3389/fmars.2020.00682 (2020).
-
Lee, K. N., Kelly, R. P., Demir-Hilton, E., Laschever, E. & Allan, E. A. Adoption of environmental DNA in public agency practice. Environ. DNA https://doi.org/10.1002/edn3.470 (2024).
-
Sander, M. et al. Environmental DNA time series analysis of a temperate stream reveals distinct seasonal community and functional shifts. River Res. Appl. 40, 850–862 (2024).
-
Tillotson, M. D. et al. Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 220, 1–11 (2018).
-
Formel, N., Enochs, I. C., Sinigalliano, C., Anderson, S. R. & Thompson, L. R. Subsurface automated samplers for eDNA (SASe) for biological monitoring and research. HardwareX 10, e00239 (2021).
-
Hendricks, A. et al. A miniaturized and automated eDNA sampler: application to a marine environment. In OCEANS 2022, Hampton Roads https://doi.org/10.1109/oceans47191.2022.9977218 (IEEE, 2022).
-
Hendricks, A. et al. Compact and automated eDNA sampler for in situ monitoring of marine environments. Sci. Rep. 13, 5210 (2023).
-
George, S. D. et al. Field trials of an autonomous eDNA sampler in lotic waters. Environ. Sci. Technol. 58, 20942–20953 (2024).
-
Preston, C. M. et al. Underwater application of quantitative PCR on an ocean mooring. PLoS ONE 6, e22522 (2011).
-
Hansen, B. K. et al. Remote, autonomous real-time monitoring of environmental DNA from commercial fish. Sci. Rep. 10, 13272 (2020).
-
Sepulveda, A. J. et al. Robotic environmental DNA bio-surveillance of freshwater health. Sci. Rep. 10, 14389 (2020).
-
Maiello, G. et al. Little samplers, big fleet: eDNA metabarcoding from commercial trawlers enhances ocean monitoring. Fish. Res. 249, 106259 (2022).
-
Chen, X. et al. Comparative evaluation of common materials as passive samplers of environmental DNA. Environ. Sci. Technol. 56, 10798–10807 (2022).
-
Pont, D. Predicting downstream transport distance of fish eDNA in lotic environments. Mol. Ecol. Resour. 24, e13934 (2024).
-
Van Driessche, C., Everts, T., Neyrinck, S. & Brys, R. Experimental assessment of downstream environmental DNA patterns under variable fish biomass and river discharge rates. Environ. DNA 5, 102–116 (2023).
-
Brantschen, J. et al. Habitat suitability models reveal the spatial signal of environmental DNA in riverine networks. Ecography https://doi.org/10.1111/ecog.07267 (2024).
-
Cantera, I. et al. Low level of anthropization linked to harsh vertebrate biodiversity declines in Amazonia. Nat. Commun. 13, 3290 (2022).
-
Zong, S. et al. Combining environmental DNA with remote sensing variables to map fish species distributions along a large river. Remote Sens. Ecol. Conserv. 10, 220–235 (2024).
-
Jeunen, G.-J. et al. Water stratification in the marine biome restricts vertical environmental DNA (eDNA) signal dispersal. Environ. DNA 2, 99–111 (2020).
-
Jeunen, G.-J. et al. Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol. Ecol. Resour. 19, 426–438 (2019).
-
Laporte, M. et al. Caged fish experiment and hydrodynamic bidimensional modeling highlight the importance to consider 2D dispersion in fluvial environmental DNA studies. Environ. DNA 2, 362–372 (2020).
-
Sansom, B. J. & Sassoubre, L. M. Environmental DNA (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environ. Sci. Technol. 51, 14244–14253 (2017).
-
Andruszkiewicz, E. A. et al. Modeling environmental DNA transport in the coastal ocean using Lagrangian particle tracking. Front. Mar. Sci. https://doi.org/10.3389/fmars.2019.00477 (2019).
-
Fukaya, K. et al. Estimating fish population abundance by integrating quantitative data on environmental DNA and hydrodynamic modelling. Mol. Ecol. 30, 3057–3067 (2021).
-
Carraro, L. & Altermatt, F. eDITH: an R�package to spatially project eDNA�based biodiversity across river networks with minimal prior information. Methods Ecol. Evol. 15, 806–815 (2024).
-
Carraro, L., Mächler, E., Wüthrich, R. & Altermatt, F. Environmental DNA allows upscaling spatial patterns of biodiversity in freshwater ecosystems. Nat. Commun. 11, 3585 (2020).
-
Blackman, R. C., Carraro, L., Keck, F. & Altermatt, F. Measuring the state of aquatic environments using eDNA-upscaling spatial resolution of biotic indices. Philos. Trans. R. Soc. Lond. B 379, 20230121 (2024).
-
Jerde, C. L. et al. Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Can. J. Fish. Aquat. Sci. 70, 522–526 (2013).
-
Rees, H. C. et al. The application of eDNA for monitoring of the great crested newt in the UK. Ecol. Evol. 4, 4023–4032 (2014).
-
Jahn, K. et al. Early detection and surveillance of SARS-CoV-2 genomic variants in wastewater using COJAC. Nat. Microbiol. 7, 1151–1160 (2022).
-
Feist, S. M. & Lance, R. F. Genetic detection of freshwater harmful algal blooms: a review focused on the use of environmental DNA (eDNA) in Microcystis aeruginosa and Prymnesium parvum. Harmful Algae 110, 102124 (2021).
-
Abdul Manaff, A. H. N. et al. Mapping harmful microalgal species by eDNA monitoring: a large-scale survey across the southwestern South China Sea. Harmful Algae 129, 102515 (2023).
-
Blackman, R. C. et al. Targeted and passive environmental DNA approaches outperform established methods for detection of quagga mussels, Dreissena rostriformis bugensis in flowing water. Ecol. Evol. 10, 13248–13259 (2020).
-
Danziger, A. M. & Frederich, M. Challenges in eDNA detection of the invasive European green crab, Carcinus maenas. Biol. Invasions 24, 1881–1894 (2022).
-
Mansfeldt, C. et al. Microbial community shifts in streams receiving treated wastewater effluent. Sci. Total Environ. 709, 135727 (2020).
-
Yamamoto, S. et al. Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 7, 40368 (2017).
-
Inoue, Y., Miyata, K., Yamane, M. & Honda, H. Environmental nucleic acid pollution: characterization of wastewater generating false positives in molecular ecological surveys. ACS ES&T Water 3, 756–764 (2023).
-
Darling, J. A., Jerde, C. L. & Sepulveda, A. J. What do you mean by false positive. Environ. DNA 3, 879–883 (2020).
-
Ficetola, G. F., Taberlet, P. & Coissac, E. How to limit false positives in environmental DNA and metabarcoding? Mol. Ecol. Resour. 16, 604–607 (2016).
-
McCauley, M., Koda, S. A., Loesgen, S. & Duffy, D. J. Multicellular species environmental DNA (eDNA) research constrained by overfocus on mitochondrial DNA. Sci. Total Environ. 912, 169550 (2024).
-
Pilliod, D. S., Goldberg, C. S., Arkle, R. S. & Waits, L. P. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 70, 1123–1130 (2013).
-
Doi, H. et al. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw. Biol. 62, 30–39 (2017).
-
Di Muri, C. et al. Read counts from environmental DNA (eDNA) metabarcoding reflect fish abundance and biomass in drained ponds. MBMG 4, e56959 (2020).
-
Pont, D. et al. Quantitative monitoring of diverse fish communities on a large scale combining eDNA metabarcoding and qPCR. Mol. Ecol. Resour. 23, 396–409 (2023).
-
Nakagawa, H., Fukushima, K., Sakai, M., Wu, L. & Minamoto, T. Relationships between the eDNA concentration obtained from metabarcoding and stream fish abundance estimated by the removal method under field conditions. Environ. DNA 4, 1369–1380 (2022).
-
Fonseca, V. G. Pitfalls in relative abundance estimation using eDNA metabarcoding. Mol. Ecol. Resour. 18, 923–926 (2018).
-
Yates, M. C., Fraser, D. J. & Derry, A. M. Meta�analysis supports further refinement of eDNA for monitoring aquatic species�specific abundance in nature. Environ. DNA 1, 5–13 (2019).
-
Sepulveda, A. J. et al. It’s complicated environmental DNA as a predictor of trout and char abundance in streams. Can. J. Fish. Aquat. Sci. 78, 422–432 (2021).
-
Sigsgaard, E. E. et al. Population characteristics of a large whale shark aggregation inferred from seawater environmental DNA. Nat. Ecol. Evol. 1, 0004 (2016).
-
Weitemier, K. et al. Estimating the genetic diversity of Pacific salmon and trout using multigene eDNA metabarcoding. Mol. Ecol. 30, 4970–4990 (2021).
-
Parsons, K. M., Everett, M., Dahlheim, M. & Park, L. Water, water everywhere: environmental DNA can unlock population structure in elusive marine species. R. Soc. Open Sci. 5, 180537 (2018).
-
Elbrecht, V., Vamos, E. E., Steinke, D. & Leese, F. Estimating intraspecific genetic diversity from community DNA metabarcoding data. PeerJ 6, e4644 (2018).
-
Turon, X., Antich, A., PalacÃn, C., Praebel, K. & Wangensteen, O. S. From metabarcoding to metaphylogeography: separating the wheat from the chaff. Ecol. Appl. 30, e02036 (2020).
-
Couton, M., Viard, F. & Altermatt, F. Opportunities and inherent limits of using environmental DNA for population genetics. Environ. DNA 5, 1048–1064 (2023).
-
Andres, K. J., Sethi, S. A., Lodge, D. M. & Andrés, J. Nuclear eDNA estimates population allele frequencies and abundance in experimental mesocosms and field samples. Mol. Ecol. 30, 685–697 (2021).
-
Wolf, K. K. E. et al. Revealing environmentally driven population dynamics of an Arctic diatom using a novel microsatellite PoolSeq barcoding approach. Environ. Microbiol. 23, 3809–3824 (2021).
-
Barbour, M. T., Gerritsen, J., Snyder, B. D. & Stribling, J. B. US Environmental Protection Agency. Rapid Bioassessment Protocols for Use in Streams and Wadeable Rivers (US EPA, 1999).
-
Cordier, T. et al. Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap. Mol. Ecol. 30, 2937–2958 (2021).
-
Yang, J. et al. Ecogenomics of zooplankton community reveals ecological threshold of ammonia nitrogen. Environ. Sci. Technol. 51, 3057–3064 (2017).
-
Nuy, J. K. et al. Responses of stream microbes to multiple anthropogenic stressors in a mesocosm study. Sci. Total Environ. 633, 1287–1301 (2018).
-
Li, F. et al. Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers. Environ. Sci. Technol. 52, 11708–11719 (2018).
-
Blackman, R. C., Ho, H.-C., Walser, J.-C. & Altermatt, F. Spatio-temporal patterns of multi-trophic biodiversity and food-web characteristics uncovered across a river catchment using environmental DNA. Commun. Biol. 5, 259 (2022).
-
Stevens, J. D. & Parsley, M. B. Environmental RNA applications and their associated gene targets for management and conservation. Environ. DNA 5, 227–239 (2023).
-
Bergsveinson, J. et al. Metatranscriptomic insights into the response of river biofilm communities to ionic and nano-zinc oxide exposures. Front. Microbiol. 11, 267 (2020).
-
Hechler, R. M., Yates, M. C., Chain, F. J. J. & Cristescu, M. E. Environmental transcriptomics under heat stress: can environmental RNA reveal changes in gene expression of aquatic organisms? Mol. Ecol. https://doi.org/10.1111/mec.17152 (2023).
-
Cordier, T. et al. Predicting the ecological quality status of marine environments from eDNA metabarcoding data using supervised machine learning. Environ. Sci. Technol. 51, 9118–9126 (2017).
-
Keck, F., Brantschen, J. & Altermatt, F. A combination of machine-learning and eDNA reveals the genetic signature of environmental change at the landscape levels. Mol. Ecol. 32, 4791–4800 (2023).
-
Salis, R. K., Bruder, A., Piggott, J. J., Summerfield, T. C. & Matthaei, C. D. High-throughput amplicon sequencing and stream benthic bacteria: identifying the best taxonomic level for multiple-stressor research. Sci. Rep. 7, 44657 (2017).
-
Sagova-Mareckova, M. et al. Expanding ecological assessment by integrating microorganisms into routine freshwater biomonitoring. Water Res. 191, 116767 (2021).
-
Cordier, T., Lanzén, A., Apothéloz-Perret-Gentil, L., Stoeck, T. & Pawlowski, J. Embracing environmental genomics and machine learning for routine biomonitoring. Trends Microbiol. 27, 387–397 (2019).
-
MartÃnez-Santos, M. et al. Treated and untreated wastewater effluents alter river sediment bacterial communities involved in nitrogen and sulphur cycling. Sci. Total Environ. 633, 1051–1061 (2018).
-
Andújar, C. et al. Metabarcoding of freshwater invertebrates to detect the effects of a pesticide spill. Mol. Ecol. 27, 146–166 (2018).
-
Vasselon, V., Rimet, F., Tapolczai, K. & Bouchez, A. Assessing ecological status with diatoms DNA metabarcoding: scaling-up on a WFD monitoring network (Mayotte island, France). Ecol. Indic. 82, 1–12 (2017).
-
Apothéloz-Perret-Gentil, L. et al. Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring. Mol. Ecol. Resour. 17, 1231–1242 (2017).
-
Feio, M. J. et al. A taxonomy-free approach based on machine learning to assess the quality of rivers with diatoms. Sci. Total Environ. 722, 137900 (2020).
-
Frühe, L. et al. Supervised machine learning is superior to indicator value inference in monitoring the environmental impacts of salmon aquaculture using eDNA metabarcodes. Mol. Ecol. 30, 2988–3006 (2021).
-
Cordier, T. et al. Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol. Ecol. Resour. 18, 1381–1391 (2018).
-
Wilkinson, S. P. et al. TICI: a taxon-independent community index for eDNA-based ecological health assessment. PeerJ 12, e16963 (2024).
-
Zhang, Y., Zhang, X., Li, F. & Altermatt, F. Fishing eDNA in one of the world’s largest rivers: a case study of cross-sectional and depth profile sampling in the Yangtze. Environ. Sci. Technol. 57, 21691–21703 (2023).
-
Gold, Z., Sprague, J., Kushner, D. J., Zerecero Marin, E. & Barber, P. H. eDNA metabarcoding as a biomonitoring tool for marine protected areas. PLoS ONE 16, e0238557 (2021).
-
Stewart, K., Ma, H., Zheng, J. & Zhao, J. Using environmental DNA to assess population-wide spatiotemporal reserve use. Conserv. Biol. 31, 1173–1182 (2017).
-
McClenaghan, B. et al. Harnessing the power of eDNA metabarcoding for the detection of deep-sea fishes. PLoS ONE 15, e0236540 (2020).
-
Fujiwara, Y. et al. Detection of the largest deep-sea-endemic teleost fish at depths of over 2,000 m through a combination of eDNA metabarcoding and baited camera observations. Front. Mar. Sci. https://doi.org/10.3389/fmars.2022.945758 (2022).
-
van der Heyde, M. et al. Taking eDNA underground: factors affecting eDNA detection of subterranean fauna in groundwater. Mol. Ecol. Resour. 23, 1257–1274 (2023).
-
Savio, D. et al. Bacterial diversity along a 2600 km river continuum. Environ. Microbiol. 17, 4994–5007 (2015).
-
De Ventura, L., Kopp, K., Seppälä, K. & Jokela, J. Tracing the quagga mussel invasion along the Rhine River system using eDNA markers: early detection and surveillance of invasive zebra and quagga mussels. MBio 8, 101–112 (2017).
-
Adams, A. J. et al. From eDNA to decisions using a multi-method approach to restoration planning in streams. Sci. Rep. 14, 14335 (2024).
-
Mahon, A. R. et al. Validation of eDNA surveillance sensitivity for detection of Asian carps in controlled and field experiments. PLoS ONE 8, e58316 (2013).
-
US Fish and Wildlife Service. Quality assurance project plan eDNA monitoring of bighead and silver carps. USFWS Great Lakes region 3 (US FWS, 2022).
-
Ellis, M. R. et al. Detecting marine pests using environmental DNA and biophysical models. Sci. Total Environ. 816, 151666 (2022).
-
Matejusova, I. et al. Environmental DNA based surveillance for the highly invasive carpet sea squirt Didemnum vexillum: a targeted single-species approach. Front. Mar. Sci. https://doi.org/10.3389/fmars.2021.728456 (2021).
-
Sepulveda, A. J. et al. When are environmental DNA early detections of invasive species actionable? J. Environ. Manage. 343, 118216 (2023).
-
Li, F. et al. Human activities’ fingerprint on multitrophic biodiversity and ecosystem functions across a major river catchment in China. Glob. Chang. Biol. 26, 6867–6879 (2020).
-
Lanzén, A., Dahlgren, T. G., Bagi, A. & Hestetun, J. T. Benthic eDNA metabarcoding provides accurate assessments of impact from oil extraction, and ecological insights. Ecol. Indic. 130, 108064 (2021).
-
Suzzi, A. L. et al. eDNA metabarcoding reveals shifts in sediment eukaryote communities in a metal contaminated estuary. Mar. Pollut. Bull. 191, 114896 (2023).
-
Stoeck, T. et al. Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture. Mar. Pollut. Bull. 127, 139–149 (2018).
-
Keck, F. et al. Meta-analysis shows both congruence and complementarity of DNA and eDNA metabarcoding to traditional methods for biological community assessment. Mol. Ecol. 31, 1820–1835 (2022).
-
Leese, F. et al. Why we need sustainable networks bridging countries, disciplines, cultures and generations for aquatic biomonitoring 2.0: a perspective derived from the DNAqua-net COST action. Adv. Ecol. Res. 58, 63–99 (2018).
-
Pont, D. et al. The future of fish-based ecological assessment of European rivers: from traditional EU Water Framework Directive compliant methods to eDNA metabarcoding-based approaches. J. Fish Biol. 98, 354–366 (2021).
-
Pawlowski, J., Bonin, A., Boyer, F., Cordier, T. & Taberlet, P. Environmental DNA for biomonitoring. Mol. Ecol. 30, 2931–2936 (2021).
-
Meyer, A. et al. Morphological vs. DNA metabarcoding approaches for the evaluation of stream ecological status with benthic invertebrates: testing different combinations of markers and strategies of data filtering. Mol. Ecol. 30, 3203–3220 (2021).
-
Blackman, R. C. et al. Advancing the use of molecular methods for routine freshwater macroinvertebrate biomonitoring — the need for calibration experiments. MBMG 3, e34735 (2019).
-
Pawlowski, J., Apothéloz-Perret-Gentil, L., Mächler, E. & Altermatt, F. Environmental DNA Applications for Biomonitoring and Bioassessment in Aquatic Ecosystems. Guidelines. Environmental Studies no. 2010: 71 (Federal Office for the Environment, 2020).
-
Laamanen, T. et al. Technology readiness level of biodiversity monitoring with molecular methods – where are we on the road to routine implementation? Metabarcoding Metagenom. 9, e130834 (2025).
-
Yang, J., Zhang, L., Mu, Y. & Zhang, X. Small changes make big progress: a more efficient eDNA monitoring method for freshwater fish. Environ. DNA 5, 363–374 (2023).
-
Shea, M. M. et al. Systematic review of marine environmental DNA metabarcoding studies: toward best practices for data usability and accessibility. PeerJ 11, e14993 (2023).
-
Li, J., Lawson Handley, L.-J., Read, D. S. & Hänfling, B. The effect of filtration method on the efficiency of environmental DNA capture and quantification via metabarcoding. Mol. Ecol. Resour. 18, 1102–1114 (2018).
-
Deiner, K. et al. Optimising the detection of marine taxonomic richness using environmental DNA metabarcoding: the effects of filter material, pore size and extraction method. MBMG 2, e28963 (2018).
-
Loeza-Quintana, T., Abbott, C. L., Heath, D. D., Bernatchez, L. & Hanner, R. H. Pathway to increase standards and competency of eDNA surveys (PISCeS)— advancing collaboration and standardization efforts in the field of eDNA. Environ. DNA 2, 255–260 (2020).
-
Altermatt, F. et al. Quantifying biodiversity using eDNA from water bodies: general principles and recommendations for sampling designs. Environ. DNA 5, 671–682 (2023).
-
ISO/DIS 17805:2023. Water Quality — Sampling, Capture and Preservation of Environmental DNA from Water (ISO, 2023).
-
Weigand, H. et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: gap-analysis and recommendations for future work. Sci. Total Environ. 678, 499–524 (2019).
-
Li, F. et al. Gap analysis for DNA-based biomonitoring of aquatic ecosystems in China. Ecol. Indic. 137, 108732 (2022).
-
Briski, E., Ghabooli, S., Bailey, S. A. & MacIsaac, H. J. Are genetic databases sufficiently populated to detect non-indigenous species? Biol. Invasions 18, 1911–1922 (2016).
-
Mc Cartney, A. M. et al. The European Reference Genome Atlas: piloting a decentralised approach to equitable biodiversity genomics. npj Biodivers. 3, 28 (2024).
-
Hebert, P. D. N., Floyd, R., Jafarpour, S. & Prosser, S. W. J. Barcode 100K specimens: in a single nanopore run. Mol. Ecol. Resour. 25, e14028 (2025).
-
Pomerantz, A. et al. Real-time DNA barcoding in a rainforest using nanopore sequencing: opportunities for rapid biodiversity assessments and local capacity building. Gigascience 7, giy033 (2018).
-
Lin, D. et al. The TRUST principles for digital repositories. Sci. Data 7, 144 (2020).
-
Leigh, D. M. et al. Best practices for genetic and genomic data archiving. Nat. Ecol. Evol. 8, 1224–1232 (2024).
-
Nilsson, R. H. et al. Introducing guidelines for publishing DNA-derived occurrence data through biodiversity data platforms. MBMG 6, e84960 (2022).
-
Berry, O. et al. Making environmental DNA (eDNA) biodiversity records globally accessible. Environ. DNA 3, 699–705 (2021).
-
Yilmaz, P. et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420 (2011).
-
Meyer, R. et al. Aligning standards communities for omics biodiversity data: sustainable Darwin Core–MIxS interoperability. Biodivers. Data J. 11, e112420 (2023).
-
Abarenkov, K. et al. Publishing DNA-derived data through biodiversity data platforms v1.3 (GBIF Secretariat, 2023).
-
Klymus, K. E. et al. The MIEM guidelines: minimum information for reporting of environmental metabarcoding data. MBMG https://doi.org/10.3897/mbmg.8.128689 (2024).
-
Takahashi, M. et al. Best practice for publishing environmental DNA (eDNA) data according to FAIR principles. Biodivers. Inf. Sci. Stand. https://doi.org/10.3897/biss.8.137742 (2024).
-
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
-
Goldberg, C. S. et al. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307 (2016).
-
Shen, E. W., Vandenberg, J. M. & Moore, A. Sensing inequity: technological solutionism, biodiversity conservation, and environmental DNA. Biosocieties 19, 501–525 (2024).
-
Carroll, S. R. et al. The CARE principles for indigenous data governance. Data Sci. J. 19, 43 (2020).
-
Secretariat of the Convention on Biological Diversity. Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity (Convention on Biological Diversity, United Nations, 2011).
-
Stammnitz, M. R., Hartman Scholz, A. & Duffy, D. J. Environmental DNA without borders: let’s embrace decentralised genomics to meet the UN’s biodiversity targets. EMBO Rep. 25, 4095–4099 (2024).
-
Handsley-Davis, M., Kowal, E., Russell, L. & Weyrich, L. S. Researchers using environmental DNA must engage ethically with Indigenous communities. Nat. Ecol. Evol. 5, 146–148 (2021).
-
Wauchope, H. S. et al. What is a unit of nature? Measurement challenges in the emerging biodiversity credit market. Proc. Biol. Sci. 291, 20242353 (2024).
-
Bhutta, U. S., Tariq, A., Farrukh, M., Raza, A. & Iqbal, M. K. Green bonds for sustainable development: review of literature on development and impact of green bonds. Technol. Forecast. Soc. Change 175, 121378 (2022).
-
Watt, R. The fantasy of carbon offsetting. Environ. Politics 30, 1069–1088 (2021).
-
Ford, H. V. et al. A technological biodiversity monitoring toolkit for biocredits. J. Appl. Ecol. 61, 2007–2019 (2024).
-
Jarman, S. N., Berry, O. & Bunce, M. The value of environmental DNA biobanking for long-term biomonitoring. Nat. Ecol. Evol. 2, 1192–1193 (2018).
Acknowledgements
Funding is from the Swiss National Science Foundation (grant nos. 31003A_173074 and 310030_197410 to F.A., and PZ00P2_202010 to L.C.). X.Z. and Y.Z. are supported by the National Key Research and Development Program of China (2022YFC32021001 and 2021YFC3201003).
Author information
Authors and Affiliations
Contributions
Overall project lead and coordination by F.A. Overall conceptualization by F.A., M.C. and R.C.B. Discussion of content by all authors. Lead writing of article by F.A., M.C. and R.C.B. Writing and lead for specific sections by F.A., M.C., L.C., F.K., L.L.-H., F.L., X.Z., Y.Z. and R.C.B. Conceptualization of figures by F.A., M.C., R.C.B., L.C. and Y.Z. Reviewing and editing of manuscript before submission by all authors.
Corresponding author
Ethics declarations
Competing interests
X.Z. directs a translation project at Nanjing University that develops apparatus for routine eDNA biomonitoring. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Biodiversity thanks David Duffy, Adam Sepulveda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
DDBJ Sequence Read Archive: https://www.ddbj.nig.ac.jp/dra/index-e.html
DEFRA DNA Center of Excellence: https://jncc.gov.uk/our-work/dna-coe/
European Nucleotide Archive: https://www.ebi.ac.uk/ena/browser/home
GBIF: https://www.gbif.org/
GEO BON: https://geobon.org/
Global Genome Biodiversity Network: https://www.tdwg.org/standards/ggbn/
International Nucleotide Sequence Database Collaboration: https://www.insdc.org/
Joint Research Center: https://joint-research-centre.ec.europa.eu/
Nagoya Protocol: https://www.cbd.int/abs/default.shtml
National eDNA Reference Centre: https://www.ecodna.org.au/nrc
OBIS: https://obis.org/
Sequence Read Archive: https://www.ncbi.nlm.nih.gov/sra
Glossary
- Amplicon sequence variant
-
Inferred unique sequence(s) derived from high-throughput sequencing after removal of erroneous sequences.
- Biological indicator
-
Taxonomic group, such as fish, macroinvertebrates or diatoms, specifically used to assess environmental conditions in relation to legislative frameworks.
- DNA barcoding
-
Identification of a specimen using a short DNA fragment called a (genetic) marker.
- High-throughput sequencing
-
Approaches used to sequence millions of DNA sequences in a rapid and cost-effective manner (also known as next-generation sequencing).
- Marker (or genetic marker)
-
A DNA sequence of a gene or part of a gene with a known location in the genome used to identify specific species.
- Metabarcoding
-
Identification of the multiple organisms represented in a sample by sequencing a common DNA marker using high-throughput sequencing.
- Operational taxonomic unit
-
An operational definition of clustered sequences based on their sequence similarity (for example, >97% similarity) to reflect approximated taxonomic units.
- Polymerase chain reaction (PCR)
-
The process used to multiply target DNA sequences in a sample to facilitate their identification.
- Primer
-
A short, single-stranded DNA sequence (~18–25 bp) used to target a region of the gene to be amplified during the polymerase chain reaction.
- Quantitative PCR (qPCR)
-
Dye-based or probe-based PCR method that allows the quantification of target DNA at each PCR amplification cycle.
- Read (amplicon read)
-
Individual sequence of base pairs (here, amplified through PCR) that corresponds to a single DNA fragment.
- Species-specific assay
-
An approach in which a single species is targeted, typically using standard, quantitative or digital PCR (as opposed to metabarcoding).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Altermatt, F., Couton, M., Carraro, L. et al. Utilizing aquatic environmental DNA to address global biodiversity targets.
Nat. Rev. Biodivers. (2025). https://doi.org/10.1038/s44358-025-00044-x
-
Accepted: 31 March 2025
-
Published: 28 April 2025
-
DOI: https://doi.org/10.1038/s44358-025-00044-x
Search
RECENT PRESS RELEASES
Related Post